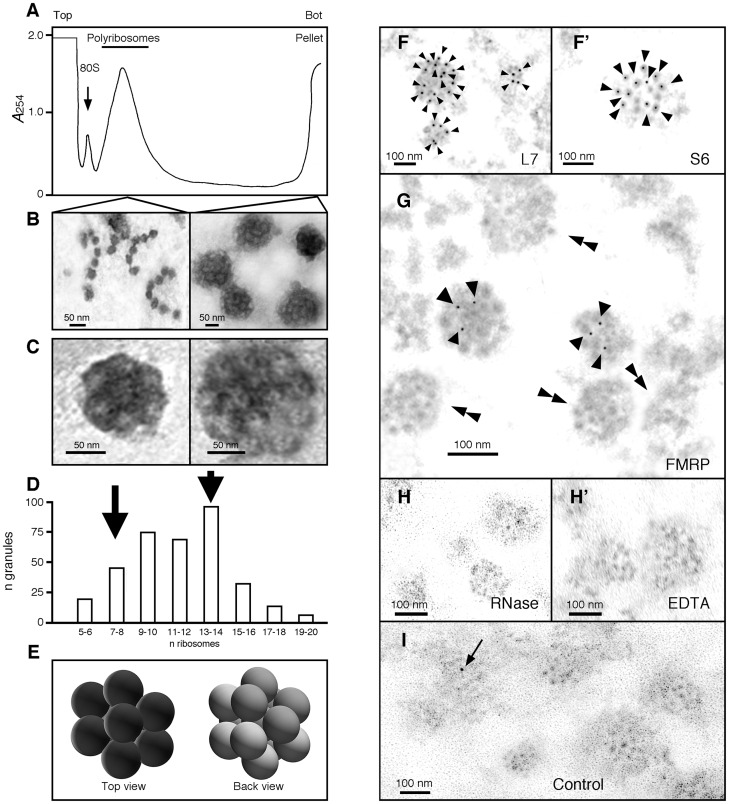
Fig 3. Ribosomes are the basic units of the granules.
A) Concentrated samples of polyribosomes were analysed by centrifugation through linear 15–60% (w/v) sucrose gradient, and fractions were collected with continuous monitoring at 254 nm. Reducing the time of centrifugation to 45 min, allowed polyribosomes to be separated from granules that sediment at the bottom of the gradient. B) Isolated polyribosomes and granules were observed by electron microscopy after negative staining. While polyribosomes present an open structure similar to beads on a string, granules display a densely compacted morula-like structure. C) Shown are two granules of two different sizes. The diameter of each unit composing the granules is similar to the reported size of 25 nm for ribosomes. D) Size distribution of granules according to their number of visible units as revealed by negative staining. Quantification of ribosomes present in each granule shows that their number varies from 5 to 20, with a mean average of 9 to14 ribosomes. E) 3D model of granules from top and back views suggests that the number of ribosomes observed in flatten EM preparations is under estimated. F and F’) Immunogold labelling of ribosomal protein L7 and S6 (15 nm, arrow heads), and G) FMRP (5 nm, arrow heads) in granules; double arrow heads point to granules free of FMRP gold signals. H and H’) RNAse and EDTA treated granules show slightly altered structures. I) Control analysis without primary antibodies showing a single contaminant signal (arrow). Ultrathin sections were obtained on materials embedded in LR-White resin.