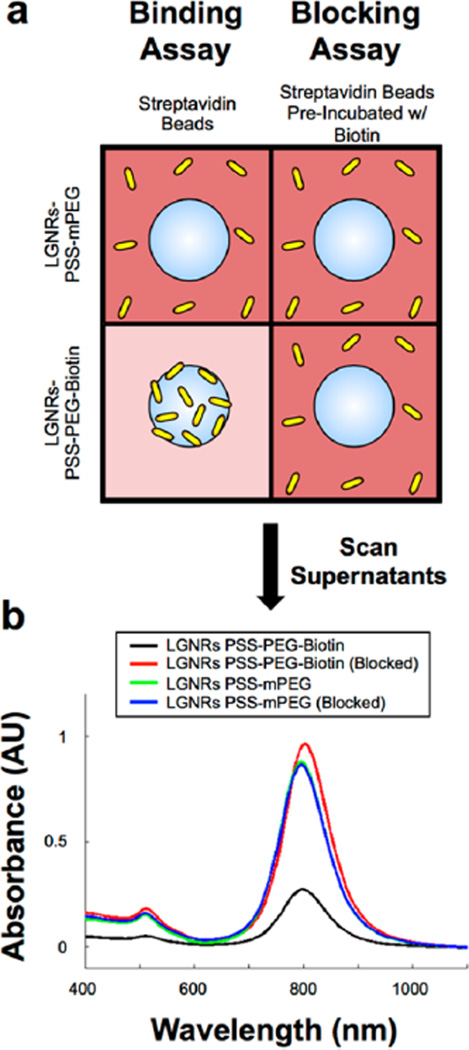
Figure 4.
LGNRs-PSS can be subsequently functionalized to achieve high-specificity molecular binding properties. (a) LGNRs-PSS-PEG-Biotin and LGNRs-PSS-mPEG were prepared and incubated with FBS to mimic biological environments. FBS-incubated GNRs were then used in biotin–streptavidin binding and blocking assays. For the binding assay, LGNRs-PSS-PEG-Biotin and LGNRs-PSS-mPEG were incubated with streptavidin-coated polystyrene beads (3 µm diameter) and centrifuged for 10s at 1000g to separate beads from free GNRs. The same process was repeated in the blocking assay, except that the streptavidin-coated beads were preincubated with excess free biotin to preclude specific binding of Large-GNRs-PSS-PEG-Biotin. (b) Absorbance measurements of the supernatant from each of the four bead-GNR combinations were taken after incubation. The same concentration of GNRs (OD 1) was used in each incubation, but only the supernatants from the incubation of LGNRs-PSS-PEG-Biotin and streptavidin beads exhibited a significant decrease in GNR concentration. These results demonstrate the proof of principle that LGNRs-PSS can be functionalized with ligands that retain molecular binding specificity in the presence of nonspecific proteins, which will be advantageous for future applications to targeted molecular imaging. Photographs from these molecular specificity assays are presented in Figure S5.