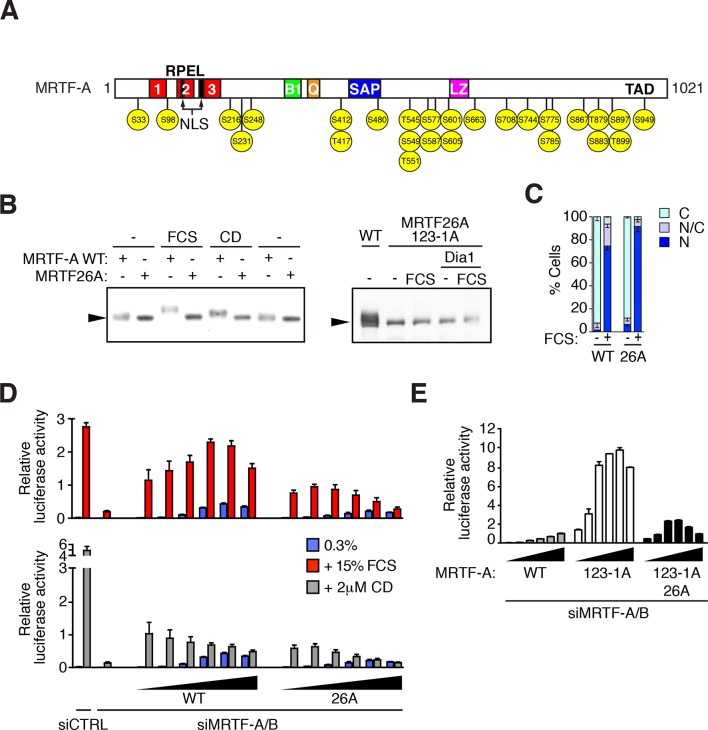
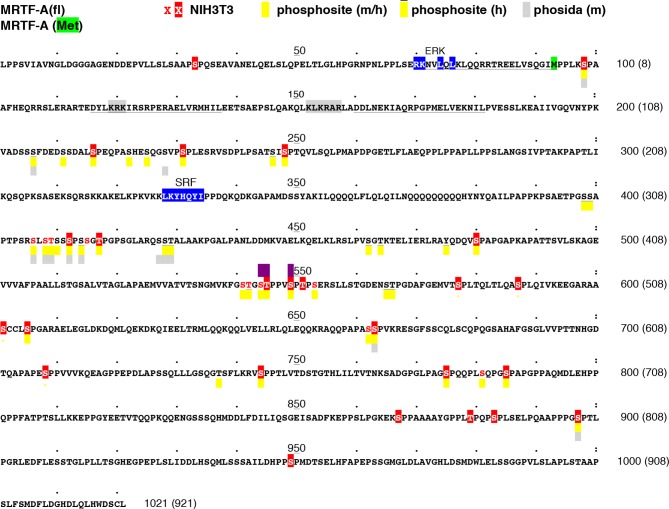
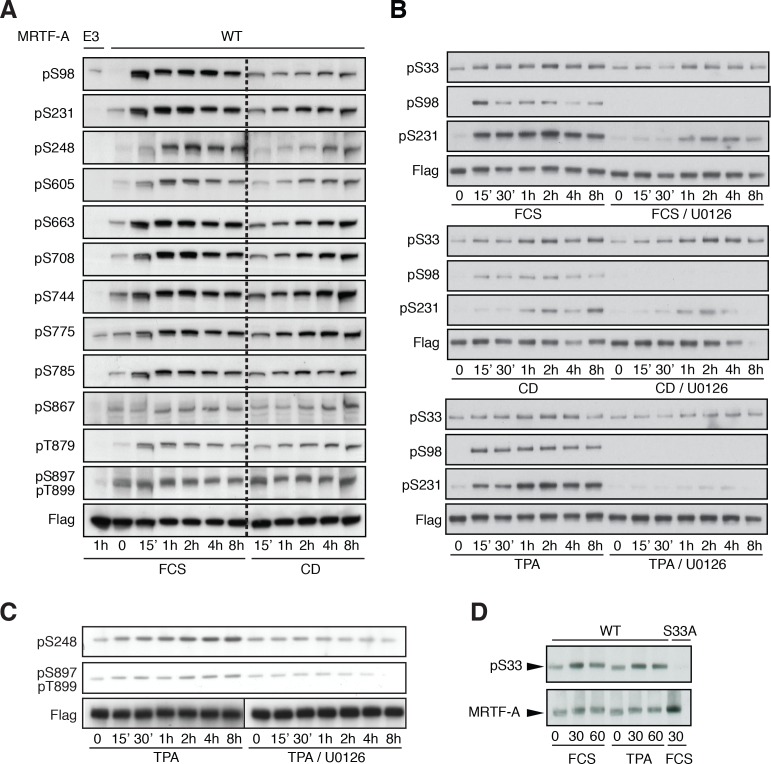
Figure 2. MRTF is phosphorylated at multiple sites.
(A) Schematic representation of MRTF-A, with S/T-P phosphorylation sites substituted with alanine in MRTF26A indicated in yellow. For details of phosphorylations, see Table 1 and Figure 2—figure supplement 1. (B) Phosphorylation is largely abolished in the MRTF26A mutant. Cells expressing wildtype MRTF-A or MRTF26A (left panel) or MRTF26A-123-1A (right panel), with or without activated mDia1, were stimulated with 15% FCS or 2 μM CD as indicated, and phosphorylation monitored by SDS-PAGE and immunoblotting. See Figure 2—figure supplement 2. (C) Cells expressing wild-type MRTF-A or MRTF-A 26A were stimulated with 15% FCS for 30 min. MRTF-A localisation was assessed by immunofluorescence (N, predominantly nuclear; N/C, pancellular; C, predominantly cytoplasmic), in two independent experiments. (D) Cells depleted of endogenous MRTFs were transfected with the MRTF-A responsive 3DA-luc SRF reporter and increasing amounts of wild-type MRTF-A or MRTF-A 26A (3, 6, 12, 25, 50, 100 ng). After treatment with 15% FCS or 2 μM CD, cell extracts were assessed for luciferase activity. Two independent experiments, data presented as mean ± half-range. (E) Cells were transfected with 3DA-luc and MRTF-A derivatives as in (D), and luciferase activity measured 24 hr later. Three independent experiments, data presented as mean ± SEM.