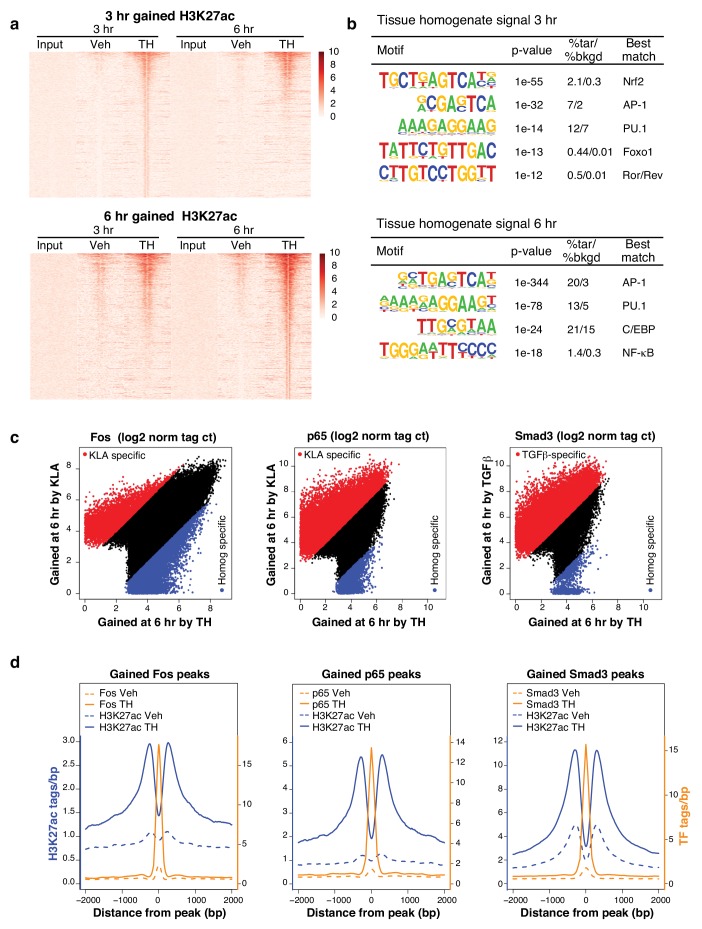
Figure 5. Complex transcriptional signals re-allocate transcription factors to novel genomic loci.
(a) H3K27ac regions differentially gained upon treatment for 3 or 6 hr with tissue homogenate compared to treatment for 3 or 6 hr with the vehicle control. The heatmap shows a 6 kb window of normalized H3K27ac tag counts of the 2510 or 5005 homogenate gained regions at 3 or 6 hr, respectively, centered on the nucleosome free region (nfr). Input shows genomic background at these regions. N = 1. (b) Motifs enriched in the vicinity of gained H3K27ac sites after treatment with tissue homogenate for 3 or 6 hr using de novo motif enrichment analysis. (c) Comparison of Fos, p65, or Smad3 tag counts at genomic regions that contain Fos, p65, or Smad3 binding after stimulation with KLA, TGFβ, or tissue homogenate. Peaks found to be differentially gained (four-fold more tags) with KLA or TGFβ are colored red, while peaks found to be differentially gained (four-fold more tags) upon tissue homogenate treatment are colored blue. N = 1. (d) Quantification of H3K27ac, Fos, p65, and Smad3 ChIP-Seq tag counts in the 6 hr vehicle or tissue homogenate treated states centered on homogenate gained (using HOMER) Fos, p65, or Smad3 binding events. Dashed lines represent ChIP-Seq signal of the vehicle state and solid lines represent the signal after 6 hr of tissue homogenate stimulation. Blue represents H3K27ac signal, orange represents signal of the respective transcription factor. N = 1.