Abstract
Objectives
To analyze the methylome of normal and osteoarthritis (OA) knee articular cartilage and determine the role of DNA methylation in the regulation of gene expression in vitro.
Methods
DNA was isolated from human normal (N=11) and OA (N=12) knee articular cartilage and analyzed by an Infinium HumanMethylation450 BeadChip methylation array. To integrate methylation and transcription, RNAseq was performed on normal and OA cartilage validated by qPCR. Functional validation was performed in the human TC28 cell line and primary chondrocytes that were treated with the DNA methylation inhibitor 5-Aza-2-deoxycytidine (5’Aza).
Results
DNA methylation profiling revealed 929 differentially methylated sites (DMS) between normal and OA cartilage that comprised a total of 500 genes. Among these, 45 transcription factors that harbored DMS were identified. Integrative analysis and subsequent validation showed a subset of 6 transcription factors that were significantly hypermethylated and downregulated in OA cartilage (ATOH8, MAFF, NCOR2, TBX4, ZBTB16 and ZHX2). Upon 5’Aza treatment, TC28 cells showed a significant increase in gene expression for all six transcription factors. In primary chondrocytes, ATOH8 and TBX4 were increased after 5’Aza treatment.
Conclusions
Our findings reveal that normal and OA knee articular cartilage have significantly different methylomes. The identification of a subset of epigenetically regulated transcription factors with reduced expression in OA may represent an important mechanism to explain changes in the chondrocyte transcriptome and function during OA pathogenesis.
Keywords: Knee OA, cartilage, DNA methylation, transcription factors, chondrocytes
INTRODUCTION
Osteoarthritis (OA) is a chronic musculoskeletal disease characterized by degradation of articular cartilage and remodeling of other tissues in synovial joints, leading to joint malfunction, pain and disability (1). It is estimated that 10–12% of adults worldwide have symptomatic OA and this number is predicted to increase 50% over the next 20 years (2). Despite the high prevalence and socioeconomic burden of the disease, OA pathophysiology is not entirely understood and, to date, there are no disease-modifying agents approved for OA treatment (3).
The pathogenesis of OA is multifactorial, with aging, obesity and genetic susceptibility as the main risk factors (4). During OA development, articular chondrocytes, the only cell type in articular cartilage that is responsible for maintaining tissue homeostasis, undergo marked transcriptional and phenotypic changes that compromise their function and lead to cartilage degradation (5). However, genome-wide association studies have shown only a small genetic variance component for OA (6) and recent evidence has pointed at epigenetic regulation as a key driver of the transcriptional alterations observed in OA chondrocytes (7–9).
Epigenetic mechanisms, defined as heritable changes in gene expression without changing the DNA sequence, include DNA methylation, post-translational histone modifications that alter chromatin structure, and complex noncoding RNA networks (10). DNA methylation consists of the addition of a methyl group to a cytosine in a cytosine-phosphate-guanine dinucleotide (CpG) to form methyl-cytosine. Genomic localization of the CpG sites is a determinant for the functional consequences of DNA methylation. Methylation in promoter and enhancer regions is known to correlate with gene silencing, whereas methylation in gene body regions usually correlates with increased gene expression (11). During development and in adulthood, DNA methylation is an essential mechanism to ensure cell-specific gene expression and, consequently, cells from different tissues often exhibit unique methylation landscapes. On the other hand, aberrant epigenetic alterations have been suggested to play a pivotal role in different pathologies, such as cancer and neurodegenerative diseases (12).
The role of DNA methylation in OA has started to be elucidated. Several candidate gene studies have identified alterations in the methylation status of genes involved in OA pathogenesis, such as MMP-3, MMP9, MMP13, ADAMTS4 (13–15), IL1B (16), NOS2 (17), GDF5 (18), SOD2 (19), and SOX9 (20). These methylation changes have been proposed to contribute to the differential gene expression observed in OA cartilage. More recently, genome wide approaches have been used to compare the methylomes of normal and OA chondrocytes (21–24). These studies clearly showed that there is a distinct methylation landscape in OA cartilage compared with healthy cartilage in hip and knee (21, 23). Moreover, they showed that hip and knee cartilage have different methylation profiles, regardless of disease status (24).
In the present study, we aimed to comprehensively compare the methylome of normal and OA knee articular cartilage using the Illumina Infinium HumanMethylation450 array. In order to identify functional changes in gene expression due to differences in methylation, we focused the second part of the study on transcription factors, proteins that bind to specific DNA sequences, thereby controlling the rate of transcription of several genes. Using integrative approaches, we identified a group of hypermethylated transcription factors with reduced expression in OA articular cartilage. We used in vitro approaches to experimentally validate the link between DNA methylation and gene expression.
METHODS
Human cartilage samples
Macroscopically normal human knee cartilage was provided by tissue banks from donors without history of previous joint pathology. OA cartilage was obtained from patients undergoing knee replacement surgery. Cartilage samples were harvested from identical locations on the weight-bearing region of the medial femoral condyle. All human cartilage samples were macroscopically assessed and scored according to the Outerbridge method (25), and histological analysis was performed on Safranin O stained sections. Based on the macroscopic and histologic analyses, samples were classified as normal or osteoarthritic. The tissue donors included in the present study are described in Supplemental Table 1.
DNA isolation and methylation profiling
DNA was extracted from snap-frozen cartilage samples using the DNeasy Plant Maxi kit (Qiagen, Valencia, CA). Resulting DNA solution was precipitated with 1.5 volumes of cold isopropanol for 30 minutes at −20°C followed by a 1-hour centrifugation at 4°C. Finally, the pellet was washed twice in 70% ethanol and resuspended in AE buffer (10 mM Tris-Cl, 0.5 mM EDTA, pH 9.0) at a concentration of 75 ng/µl. Then, 300 ng of DNA were bisulfite treated using the EZ DNA methylation kit (Zymo Research Corp., Orange, CA) and bisulfite-converted DNA was used to hybridize Infinium HumanMethylation450 BeadChips (Illumina, San Diego, CA).
DNA methylation data analysis
Raw intensity data was processed with the Bioconductor package Minfi (26) and normalized for technical variations using SWAN (27). Poorly performing probes (P>0.01 in more than one sample) and probes that interrogated high frequency single nucleotide polymorphisms or mapped on sex chromosomes were removed from the analysis. The final dataset included 446950 probes. The β value, which represents the percentage of methylation at each CpG site, was calculated from the normalized intensity values. β values at all CpG sites were compared between OA and normal samples using an F-test after including age, BMI and sex as covariates and P-values were corrected for multiple comparisons following the Benjamini-Hochberg procedure. Differentially methylated CpG sites (DMS) were defined as having Benjamini-Hochberg adjusted p-values < 0.05 and a difference in mean β values > 0.15 between normal and OA groups. To define differentially methylated genes (DMG), the methylation data was mapped to the genome using Illumina’s v1.2 annotation from the Bioconductor package IlluminaHumanMethylation450kanno.ilmn12.hg19, which is based on UCSC hg19 (NCBI Reference Sequence Database Release 37).
Gene Ontology (GO) enrichment analyses were performed using the web-based gene set analysis toolkit WebGestalt. Pathway analysis was performed for differentially methylated genes with the Bioconductor package SPIA using topologies derived from the KEGG database. In both cases, a hypergeometric test was used to test for enrichment after multiple testing adjustment with the Benjamini-Hochberg procedure. An adjusted p-value < 0.05 was considered significant.
Enrichment analysis for different genomic regions was performed by comparing the percentage of differentially methylated sites (DM rate) with the total number of sites present in the array after filtering (Assay rate). Statistical analysis applied the Fischer test with multiple testing adjustment with the Benjamini-Hochberg procedure. A corrected p-value < 0.05 was considered significant.
An outline of the analyses used in the present study is shown in Supplementary Figure 1.
RNA isolation and RNA-seq
Genome-wide transcriptomic analysis was performed in a set of 8 normal and 10 OA knee articular cartilage samples. RNA was isolated from cartilage stored at −20°C in Allprotect Tissue Reagent (Qiagen, Valencia, CA). Briefly, cartilage was pulverized using a 6770 Freezer/Mill Cryogenic Grinder (SPEX SamplePrep, Metuchen, NJ), and homogenized in Qiazol Lysis Reagent (Qiagen, Valencia, CA) at a concentration of 25mg tissue sample per 700ul Qiazol. Then, RNA was isolated using the mRNeasy Mini kit with on-column DNAse digestion according to the manufacturer’s protocol, followed by decontamination of proteoglycans using RNAmate (BioChain Institute, Newark, CA). RNA purity was assessed using NanoDrop (ND-1000, Thermo Scientific, Wilmington, DE) and sample quality was determined with the Agilent 2100 Bioanalyzer using the RNA 6000 Nano LabChip (Agilent 5067-1511, Santa Clara, CA). Normal and OA cartilage samples had an RNA Integrity Number (RIN) of 3.9±1.1 and 5.6±1.7, respectively. Sequencing mRNA libraries were prepared using the Encore Complete RNA-Seq DR Multiplex System 1–8 and 9–16 (NuGen, San Carlos, CA). Libraries were amplified by PCR, gel purified, and Illumina HiSeq 2000 was used to generate a total of 8–30 million 100bp reads.
RNA-seq reads for each library were mapped to the UCSC human reference genome hg19 downloaded from the Illumina iGenome website (http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn) using TopHat2 (v2.0.9)18 and Bowtie2 (v2.1.0)19 with the default settings and options. The aligned reads were assembled into transcripts and their abundances were estimated using Cufflinks (v2.1.1). Transcript abundances were measured in Fragments Per Kilobase of exon per Million fragments mapped (FPKM), which describes the relative abundances of transcripts in an experiment after normalization to the transcript size and the total number of aligned reads in the sample. Genes with a Benjamini-Hochberg adjusted p-value < 0.05 were considered significantly differentially expressed.
Quantitative polymerase chain reaction (qPCR)
Validation of mRNA expression of selected transcription factors as detected by RNA-seq was performed in an independent set of 13 normal and 11 OA knee articular cartilage samples. Individual gene expression was assessed by qPCR using a LightCycler 480 instrument (Roche Diagnostics) and TaqMan gene expression assay probes (Applied Biosciences) for ATOH8, FOXO3, KLF15, MAFF, NCOR2, NFATC2, RARA, TBX4, ZBTB16, ZHX2 and GAPDH as described elsewhere (28). Statistical differences in qPCR experiments were determined by the Student t-test. A p-value < 0.05 was considered significant.
Cell culture
Primary human articular chondrocytes were isolated from healthy joints as previously described (28). After isolation, chondrocytes were plated at a density of 104 cells/cm2 in Dulbecco’s modified Eagle’s medium (DMEM; Fischer Scientific) containing 10% calf serum (CS) and incubated at 37°C in 5% CO2. First-passage chondrocytes were used in all experiments. The immortalized human chondrocyte cell line TC28 (29) was cultured in DMEM containing 10% CS and only cells that had been maintained for fewer than 20 passages were used in all experiments.
5-Aza-2-deoxycytidine (5’Aza) experiments followed different methodologies depending on the cell type used, due to difference in the rate of proliferation. For primary chondrocytes, freshly plated cells were incubated with or without 10µM 5’Aza for at least 3 population doublings (approximately 4–5 weeks in culture). TC28, cells were treated with 0, 1, 5 or 10 µM 5’Aza for 48 hours. At the end of the treatment, RNA was collected using Direct-Zol RNA miniprep kit (Zymo Research) and gene expression was assessed by qPCR as described above.
RESULTS
General description of normal and OA cartilage methylomes
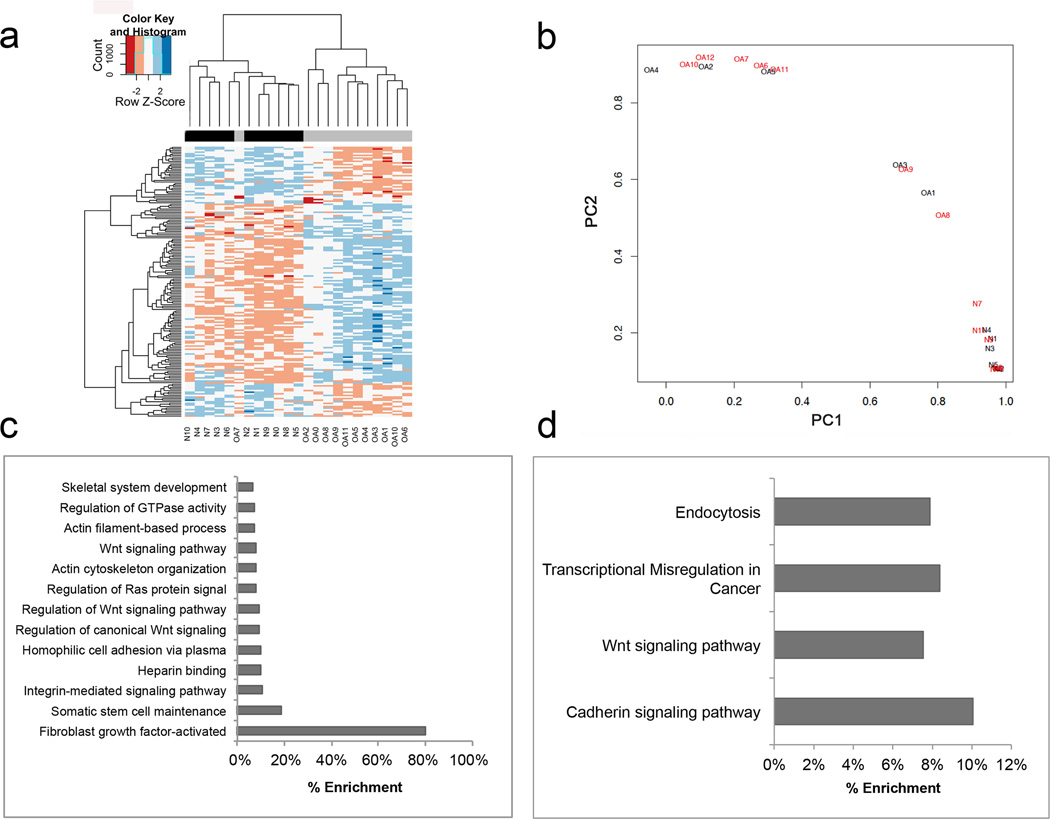
Genome-wide DNA methylation profiling was performed in 11 macroscopically normal knee cartilage samples obtained from healthy donors and 12 samples obtained from patients with OA. Unsupervised hierarchical clustering revealed that healthy and OA samples segregated in two different clusters (Figure 1a). The first cluster consisted of samples from ten normal and one OA patient, and the second cluster consisted of samples from one healthy control and eleven patients with OA.
FIGURE 1.
Genome-wide methylation analysis of human normal and OA knee articular cartilage. a) Unsupervised hierarchical clustering of DNA methylation values in 11 normal samples and 12 OA samples. b) Principal component analysis (PCA) of DNA methylation in the 929 differentially methylated CpG sites (DMS) between normal (black) and OA (red) samples. c) Gene ontology (GO) and d) Pathway analysis of the 929 DMS between normal and OA samples.
Identification and characterization of DMS in knee articular cartilage from control and OA patients
We identified a total of 929 DMS between normal and OA cartilage that comprised a total of 500 individual genes. (For a complete list of DMS see Supplemental Table 2.) Among these, 356 (38.3%) sites were hypomethylated and 573 (61.7%) hypermethylated in OA cartilage. These differences were observed after performing statistical analysis with correction by sex and BMI. However, when the analysis was corrected for age as a covariate, no DMS were observed.
Principal component analysis using the 929 DMS revealed complete segregation of normal and OA samples (Figure 1b). The gene-associated CpGs with the highest differences in methylation between both groups are shown in Table 1. In addition to the differences in the magnitude of single CpG methylation, we found several genes that harbored multiple DMS that were hypermethylated, such as LRP5, NCOR2, NFATC1, PRDM8 and TBX4, or hypomethylated in OA cartilage, such as RUNX1.
Table 1.
Most differentially hypomethylated and hypermethylated CpG sites in knee articular cartilage from normal donors and osteoarthritis patients.
| Illumina probe ID | Associated gene symbol |
Δβ (Normal-OA) |
P-value | Corrected P-value |
|---|---|---|---|---|
| Hypomethylated in OA | ||||
| cg21465150 | TRPV3 | −0.45 | 1.96E-04 | 0.041057162 |
| cg07371504 | TRPV3 | −0.43 | 2.18E-04 | 0.043347392 |
| cg20092122 | BST2 | −0.40 | 1.26E-06 | 0.004419322 |
| cg06880930 | CPNE2 | −0.34 | 3.06E-06 | 0.006008504 |
| cg13030790 | RUNX1 | −0.33 | 2.35E-05 | 0.015638525 |
| cg15591803 | DENND2D | −0.31 | 1.32E-07 | 0.00200339 |
| cg01675238 | JMJD7 | −0.31 | 4.34E-06 | 0.00688781 |
| cg22714290 | LOC148696 | −0.30 | 5.86E-06 | 0.007902923 |
| cg09966895 | ODZ4 | −0.30 | 6.13E-07 | 0.003304806 |
| cg12273284 | CAMK1D | −0.29 | 2.73E-05 | 0.016527922 |
| cg03744842 | CCM2 | −0.29 | 3.31E-06 | 0.006090967 |
| cg11854227 | ILDR1 | −0.28 | 6.16E-07 | 0.003250758 |
| cg20054157 | SFTA1P | −0.27 | 1.05E-06 | 0.004161572 |
| cg00694560 | ILDR1 | −0.27 | 3.87E-10 | 6.27085E-05 |
| cg21108085 | CD82 | −0.26 | 1.39E-08 | 0.000615655 |
| cg25758828 | PAX8 | −0.26 | 5.12E-05 | 0.021732678 |
| cg16707506 | CCM2 | −0.26 | 1.04E-05 | 0.01049113 |
| cg07168232 | LAMB3 | −0.26 | 8.71E-06 | 0.009589979 |
| cg19919590 | LAPTM5 | −0.25 | 9.11E-06 | 0.009808833 |
| cg03357727 | ULK4 | −0.25 | 4.42E-06 | 0.006881195 |
| Hypermethylated in OA | ||||
| cg03358468 | KDM4B | 0.44 | 3.22E-07 | 0.002562628 |
| cg18536148 | TBX4 | 0.39 | 2.60E-06 | 0.005864998 |
| cg15517343 | NFATC1 | 0.34 | 2.79E-07 | 0.00260294 |
| cg14196395 | DYSF | 0.33 | 1.94E-05 | 0.014468514 |
| cg01789499 | PRDM8 | 0.33 | 9.45E-06 | 0.009995321 |
| cg10890644 | TUBAL3 | 0.32 | 1.10E-04 | 0.031613525 |
| cg26819718 | C10orf11 | 0.32 | 7.99E-05 | 0.027232593 |
| cg24274579 | TBX4 | 0.30 | 1.36E-06 | 0.004576905 |
| cg03311556 | PRDM8 | 0.30 | 3.07E-06 | 0.005983571 |
| cg22079102 | KDM4B | 0.30 | 2.90E-06 | 0.005832481 |
| cg13738327 | LRP5 | 0.30 | 6.44E-05 | 0.024342721 |
| cg14419393 | ASPSCR1 | 0.30 | 9.20E-05 | 0.028974493 |
| cg06365535 | TBX4 | 0.30 | 1.19E-07 | 0.001863844 |
| cg06440348 | PRDM8 | 0.29 | 1.61E-06 | 0.004952817 |
| cg01851968 | VASN | 0.29 | 9.97E-05 | 0.03017708 |
| cg22902505 | PRDM8 | 0.29 | 1.15E-06 | 0.00431253 |
| cg07727358 | FGFRL1 | 0.29 | 1.86E-05 | 0.01429792 |
| cg14619259 | SPRY4 | 0.29 | 4.73E-05 | 0.021123812 |
| cg16536399 | NFATC1 | 0.28 | 2.99E-05 | 0.017365227 |
| cg06458239 | ZNF549 | 0.28 | 5.73E-08 | 0.001210523 |
As shown in Figure 1c, gene ontology (GO) term analysis of genes containing DMS showed significant overrepresentation of genes involved in different signaling pathways such as integrins (q<0.01), Wnt (q<0.001) and FGF (q<0.01) (see Supplemental Table 3 for complete list).
We next carried out a pathway analysis using the KEGG database for all genes containing DMS. The differentially enriched canonical pathways identified were endocytosis (q<0.05), transcriptional misregulation in cancer (q<0.05), Wnt signaling pathway (q<0.01), and cadherin signaling pathway (q<0.01) (Figure 1d).
DMS enrichment analysis for CpG genomic features
Since recent studies suggest that DNA methylation effects on gene expression are highly dependent on the CpG genomic location (11), we performed an enrichment analysis of all the DMS identified in this study for their position in a gene or relative to a CpG island. Compared with all sites included in the 450k methylation array, DMS were significantly enriched in intergenic and low density CpG regions (Table 2). In the intragenic regions, DMS were enriched in the gene body and depleted in promoters. In addition, 52% of the DMS were in regions predicted to be functional enhancers as opposed to only 21% of all the CpGs included in the methylation array, indicating that DMS are significantly (p<0.001) enriched in enhancers.
Table 2.
Enrichment analysis of differentially methylated (DM) sites for genomic features.
| Genomic region |
#DM sites | DM Rate |
#Assay sites (after filtering) |
Assay Rate (after filtering) |
Corrected P-value |
Status |
|---|---|---|---|---|---|---|
| Enhancer | 487 | 52% | 93053 | 21% | 2.20E-16 | Enriched |
| No enhancer | 442 | 48% | 353903 | 79% | 2.20E-16 | Depleted |
| Island | 367 | 40% | 287941 | 64% | 2.20E-16 | Depleted |
| No island | 562 | 60% | 159015 | 36% | 2.20E-16 | Enriched |
| Genic | 653 | 70% | 336801 | 75% | 4.50E-04 | Depleted |
| Intergenic | 276 | 30% | 110155 | 25% | 4.50E-04 | Enriched |
| Position within a gene | ||||||
| TSS1500 | 55 | 6% | 62471 | 14% | 6.27E-15 | Depleted |
| TSS200 | 28 | 3% | 48681 | 11% | 2.20E-16 | Depleted |
| 5’UTR | 91 | 10% | 38707 | 9% | N.S | |
| 1st Exon | 18 | 2% | 21436 | 5% | 7.03E-06 | Depleted |
| Body | 418 | 45% | 149763 | 34% | 4.62E-13 | Enriched |
| 3’UTR | 43 | 5% | 15743 | 4% | N.S | |
| Position within a CpG island | ||||||
| N_Shore | 103 | 11% | 57319 | 13% | N.S | |
| N_Shelf | 48 | 5% | 22449 | 5% | N.S | |
| Island | 94 | 10% | 143319 | 32% | 2.20E-16 | Depleted |
| S_Shore | 81 | 9% | 44754 | 10% | N.S | |
| S_Shelf | 41 | 4% | 20100 | 4% | N.S | |
Integrative analysis of DNA methylation and gene expression of transcription factors in normal and OA articular cartilage
To increase our functional understanding of the methylation changes observed in OA cartilage, we decided to focus on transcription factors since they act as master regulators of gene expression in eukaryotic cells. As shown in Supplemental Table 4, we identified 45 transcription factors that harbored at least one DMS. Of these differentially methylated transcription factors (DMTFs), 31 were hypermethylated and 14 hypomethylated. Whereas some of the DMTFs, such as RUNX2, NFATC1 and NCOR2, had been previously associated with OA pathology (30–32), the majority of the DMTFs have not been studied in the context of cartilage biology or OA development.
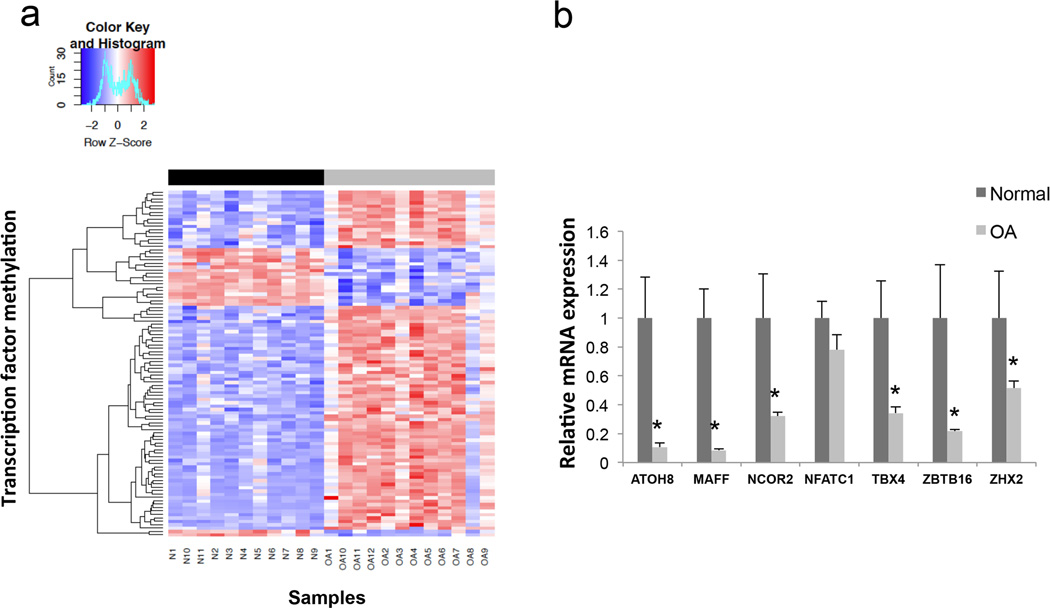
Next, we compared the methylation data and the gene expression data to determine whether these DMTFs were also differentially transcribed. As illustrated in Table 3, only 10 DMTFs overlapped between both data sets. Of these, 7 were hypermethylated in OA cartilage whereas 3 were hypomethylated. Based on the CpG methylation status of these 10 DMTFs, the hierarchical clustering analysis was able to separate healthy and OA donors (Figure 2a). Interestingly, all the DMTF that were also differentially expressed were transcriptionally repressed in OA cartilage.
Table 3.
Differentially expressed transcription factors harboring differentially methylated CpG in knee articular cartilage from normal and osteoarthritis patients.
| Methylation Status in OA |
Associated gene symbol |
CpG genomic features |
Gene expression status in OA |
Average FPKM Normal |
Average FPKM OA |
Log2 (Fold expression) |
|---|---|---|---|---|---|---|
| Hypomethylated | BCL6 | 5’UTR | Downregulated | 268.837 | 113.734 | −1.24107 |
| RORA | Body | Downregulated | 22.9409 | 10.1887 | −1.17095 | |
| ZNF395 | Body | Downregulated | 57.0915 | 16.3765 | −1.80165 | |
| Hypermethylated | ATOH8 | Body, enhancer | Downregulated | 51.8614 | 6.58004 | −2.97849 |
| MAFF | Body | Downregulated | 26.6943 | 4.13804 | −2.68951 | |
| NCOR2 | 5’UTR, body, enhancer | Downregulated | 57.9958 | 24.1427 | −1.26436 | |
| NFATC1 | Body | Downregulated | 49.741 | 22.6369 | −1.13576 | |
| TBX4 | TS200, TS500, 5’UTR, body, 3’UTR, enhancer | Downregulated | 32.8472 | 13.4769 | −1.28528 | |
| ZBTB16 | Body | Downregulated | 36.2553 | 11.9678 | −1.59903 | |
| ZHX2 | 5’UTR, enhancer | Downregulated | 23.4487 | 9.02882 | −1.3769 | |
FIGURE 2.
a) Heatmap of 102 differentially methylated CpG sites located in transcription factor genes with differential methylation among normal and OA knee articular cartilage samples. Rows represent CpG sites and columns represent samples. Dendogram at the top shows the clustering of the normal (black) and OA (grey) samples. b) Relative mRNA expression of selected transcription factors in knee articular cartilage of 13 normal and 11 OA patients, assessed by real-time polymerase chain reaction. * = P < 0.05.
To investigate the putative functionality of the observed link between methylation and transcription, we selected those DMTFs with a negative association between methylation and transcription for further study since they are more likely to be epigenetically regulated in terms of gene transcription (11). These included ATOH8, MAFF, NCOR2, NFATC1, TBX4, ZBTB16 and ZHX2.
We first confirmed differential mRNA expression by qPCR in an independent cohort of patients. As indicated in Figure 2b, a significant reduction in mRNA levels was observed for ATOH8, MAFF, NCOR2, TBX4, ZBTB16 and ZHX2 in OA samples when compared with controls. Since no differences were found for NFATC1 expression levels, this gene was excluded from subsequent experiments.
Changes in gene expression following DNA demethylation in cultured articular chondrocytes
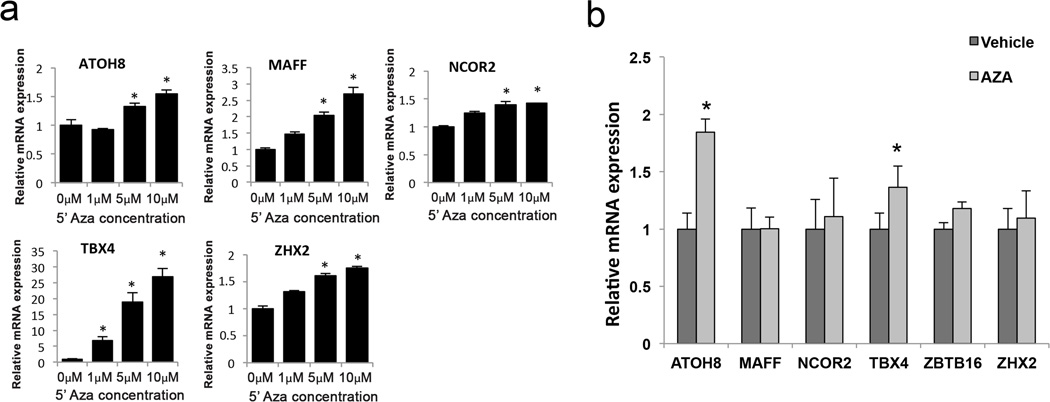
One of the limitations of previous studies is that the consequences of differential methylation on transcriptional regulation were not validated in an experimental setting. For that purpose, we tested the effects of the DNA methylation inhibitor 5’Aza on the gene expression of ATOH8, MAFF, NCOR2, TBX4, ZBTB16 and ZHX2 in cultured human articular chondrocytes. We first treated TC28 cells with increasing 5’Aza concentrations for 48 hours. As shown in Figure 3a, there was a significant dose dependent increase in mRNA levels for all the genes tested, except for ZBTB16, which was not expressed in any condition. TBX4 showed the most striking differences in expression being undetectable under basal conditions and strongly upregulated by 5’Aza treatment.
FIGURE 3.
a) Relative mRNA expression of selected transcription factors in cultured immortalized human chondrocytes (TC28) treated with different doses of the DNA methylation inhibitor 5-Aza-2-deoxycytidine (5’Aza) for 48 hours. Values are expressed as mean ± SEM. * = P < 0.05 vs 0µM 5’Aza. b) Relative mRNA expression of selected transcription factors in cultured primary human chondrocytes from normal donors (N=5) treated with 5’Aza (10µM) or vehicle for 4 weeks. Values are expressed as mean ± SEM. * = P < 0.05.
Primary human articular chondrocytes isolated from healthy donors, were treated with 5’Aza for 4–5 weeks to ensure an appropriate reduction in methylation since these cells divide very slowly and 5’Aza requires active cell division in order to effectively demethylate the DNA. As opposed to TC28 cells, primary articular chondrocytes expressed detectable levels of all transcription factors assessed in this experiment. Following 5’Aza treatment, ATOH8 and TBX4 showed significantly increased expression levels (Figure 3b).
DISCUSSION
In the present study, we compared for the first time the DNA methylation profile of knee articular cartilage from 11 normal and 12 OA donors using the most comprehensive methylation array available, the Illumina Infinium HumanMethylation450 array. We identified 929 DMS that are associated with 500 unique genes (Supplemental Table 2), revealing that healthy and OA knee articular cartilage exhibit substantially different methylomes. Furthermore, comparative analysis of methylation and gene expression allowed us to identify several hypermethylated transcription factors with reduced expression in OA cartilage.
The current report greatly expands the findings of Fernandez-Tajes et al (21) that observed 91 DMS when comparing knee cartilage samples from normal and OA patients using the Illumina Infinium HumanMethylation27 array, thus offering a broader insight on the epigenetic changes occurring during knee OA pathogenesis. We confirmed the methylation status of the most hypomethylated genes reported in that study, such as RUNX1 and KRT80, as well as some of the hypermethylated genes, including IGF2AS, SOCS1, and TBX4. However, there is a substantial difference in the number of DMS found in both studies. This could be explained by differences in the array size and design, since the HumanMethylation27 array that includes 27000 CpG sites predominantly located in gene promoters whereas the HumanMethylation450 array contains 480000 probes within a broad range of genomic features, such as enhancers, promoters, UTRs and gene bodies. In support of this notion, our enrichment analysis for specific genomic regions showed that DMS are significantly depleted in promoters, particularly in those containing CpG islands (Table 2). This observation is in concordance with the fact that epigenetic modifications are more likely to occur in CpG-poor genomic regions, such as enhancers (33, 34). Enhancer methylation is an active field of research and it has been shown to negatively correlate with transcription factor binding, enhancer activity and gene transcription (34–36). In this regard, we found significant accumulation of DMS in enhancer regions, both inter and intragenic, and similar findings have been reported in hip articular cartilage (22, 23), pointing at the possibility that specific genomic regions are more prone to undergo epigenetic changes during OA development.
An important finding initially reported by Fernandez-Tajes et al (21) and confirmed in more recent studies (22, 23, 37) is the characterization of a cluster of OA patients with a specific methylation signature enriched for inflammatory genes. While corroborating these findings in our study would have been of interest, a limitation of our study is that we did not have an adequate sample number to perform this kind of analysis. It should be also pointed out that DNA methylation can be influenced by age, sex and BMI (38). The differences in DNA methylation observed in the present study were not due to sex or BMI as covariates. However, when the data was corrected for age we did not observe any significant differences in CpG methylation. This could be due to the age difference between control and OA samples as well as to the relatively low number of samples used in the study. Therefore, a limitation of our study is that the differential DNA methylation reported can be driven by both OA and aging, and caution should be taken when interpreting these results in the context of OA. Further studies on larger numbers of young normal, OA and age matched old normal tissues are needed to identify age-related versus OA- associated changes in articular cartilage methylome.
In recent reports, hip and knee articular cartilage showed inherent differences in DNA methylation, suggesting a joint specific epigenetic landscape (23, 24). In fact, den Hollander and colleagues (24) reported differential clustering of hip and knee samples independently of disease status, prompting the notion that knee and hip OA have similar pathology but they are driven by different epigenetic mechanisms. However, we have found some commonalities when comparing our results with those reported in the above-mentioned studies. For example, a vast portion of the most differentially methylated genes (Supplementary Table 5) and, in particular, the DMTF identified in our study (Supplementary Table 6) were also differentially methylated when comparing hip OA and disease free samples (23). We therefore cannot rule out the possibility that, despite having substantial differences in the methylation landscape, knee and hip articular chondrocytes share key common epigenetically controlled genes that may play a pivotal role in tissue homeostasis and OA pathogenesis.
Genome wide expression studies have shown that aberrant gene transcription in articular chondrocytes is a hallmark of OA (39, 40). It has been repeatedly reported that several catabolic proteins, such as extracellular matrix proteases or proinflammatory factors, have increased expression in OA cartilage, whereas key anabolic genes and components of the autophagic machinery are repressed (reviewed in (41, 42)). These transcriptional changes are thought to ultimately compromise chondrocyte ability to sustain cartilage homeostasis. However, the molecular mechanisms driving these transcriptional alterations remain elusive. Recent reports identified specific transcription factors that coordinate broad cellular processes during cartilage development and OA pathogenesis. For instance, hypoxia induced factor 2 alpha (HIF2α) has been proposed as a key driver of the catabolic response observed during OA development by inducing the expression of several catabolic genes (43). On the other hand, SOX9 is a pivotal factor of chondrogenesis, inducing the expression of different chondrocyte differentiation markers, and that is strongly repressed during OA development (44, 45). Nevertheless, identification of key transcription factors, and the mechanisms controlling their expression, is paramount for the development of new therapeutic strategies to treat OA. In the present study, using an integrative approach, we identified several transcription factors that are differentially methylated and repressed in OA cartilage (Table 3). Moreover, we experimentally validated that modulation of DNA methylation is able to regulate their transcription in cultured human articular chondrocytes. Several of the transcription factors identified have not been studied in the context of OA pathophysiology, thus opening new opportunities to future studies aimed to characterize their function in articular cartilage and OA development. TBX4 is a member of a phylogenetically conserved family of transcription factors involved in the regulation of developmental processes. TBX4 regulates limb development and is required for muscle and tendon morphogenesis (46). In addition, mutations in TBX4 cause small patella syndrome in humans (47). ZBTB16, also known as ZNF145, enhanced the expression of SOX9, suggesting that ZNF145 acts as a factor upstream of SOX9, the master regulator of chondrogenesis (48).
Methylation is a dynamic process that has been linked to gene silencing. Despite active research in the field, it still remains unclear whether methylation leads to gene silencing or gene silencing precedes DNA methylation (11). In human chondrocytes, there is mounting evidence of specific genes transcriptionally controlled by epigenetic mechanisms, including SOX9 (20), GDF5 (49), MMP13 (15), and IL1B (16). The mechanisms controlling site targeted methylation and demethylation in cartilage are not known. Generally, addition of methyl groups to cytosines is catalyzed by the DNA methyl transferase (DNMT) family of proteins that is comprised of three members: DNMT1, DNMT3a, and DNMT3b. Whereas decreased DNMT1 expression and activity has been found during aging (50), no significant changes on any DNMT were found in articular cartilage from normal and OA donors (16, 51). On the other hand, the family of ten-eleven translocases (TET) mediates the addition of hydroxyl group to a methyl-cytosine to form 5-hydroxymethylcytosine, eventually leading to active demethylation through different mechanisms (52). Reduced TET1 expression and altered 5-hydroxymethylcytosine levels have been reported in OA articular cartilage in a process that is partially mediated by pro-inflammatory cytokines (53, 54). In this regard, it is important to point out that the methodology used in the present study and other recent reports (21–24, 37) does not allow us to discern between cytosine methylation and hydroxymethylation. Future studies should be aimed at unraveling the precise molecular mechanisms underlying the profound changes in methylation and hydroxymethylation observed in OA articular cartilage.
In summary, our results show that normal and OA knee articular cartilage can be distinguished by their DNA methylation profiles. We identified a number of differentially methylated CpG sites and we provided the first description of differential methylation among transcription factors in normal and OA cartilage. Furthermore, we identified a number of hypermethylated transcription factors with reduced expression in OA cartilage and experimentally validated the epigenetic control of their gene expression in human chondrocytes. These findings suggest that methylation-related changes in several important transcription factors represent an important mechanism that may explain changes in chondrocyte transcriptome and function in OA.
Supplementary Material
Overview of the design and data analysis used in the study.
Acknowledgments
We gratefully acknowledge Shantanu Patil MD, Stuart Duffy, Merissa Olmer, and Lilo Creighton for their technical assistance.
This study was supported by NIH grants AG007996, AG049617 and TR001114.
Footnotes
The authors have no conflicts of interest.
Author contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Lotz had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Alvarez-Garcia, Sasho, Lotz.
Acquisition of data. Alvarez-Garcia, Akagi, Saito.
Analysis and interpretation of data. Alvarez-Garcia, Fisch, Wineinger, Su, Lotz.
REFERENCES
- 1.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64(6):1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nature reviews Rheumatology. 2014 doi: 10.1038/nrrheum.2014.44. [DOI] [PubMed] [Google Scholar]
- 3.Hochberg MC, Altman RD, April KT, Benkhalti M, Guyatt G, McGowan J, et al. American College of Rheumatology 2012 recommendations for the use of nonpharmacologic and pharmacologic therapies in osteoarthritis of the hand, hip, and knee. Arthritis Care Res (Hoboken) 2012;64(4):465–474. doi: 10.1002/acr.21596. [DOI] [PubMed] [Google Scholar]
- 4.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best practice & research Clinical rheumatology. 2014;28(1):5–15. doi: 10.1016/j.berh.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Lotz M, Loeser RF. Effects of aging on articular cartilage homeostasis. Bone. 2012;51(2):241–248. doi: 10.1016/j.bone.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandell LJ. Etiology of osteoarthritis: genetics and synovial joint development. Nature reviews Rheumatology. 2012;8(2):77–89. doi: 10.1038/nrrheum.2011.199. [DOI] [PubMed] [Google Scholar]
- 7.Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Current rheumatology reports. 2013;15(9):353. doi: 10.1007/s11926-013-0353-z. [DOI] [PubMed] [Google Scholar]
- 8.Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18(2):109–118. doi: 10.1016/j.molmed.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Im GI, Choi YJ. Epigenetics in osteoarthritis and its implication for future therapeutics. Expert Opin Biol Ther. 2013;13(5):713–721. doi: 10.1517/14712598.2013.764410. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 12.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22(1):50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52(10):3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 14.Cheung KS, Hashimoto K, Yamada N, Roach HI. Expression of ADAMTS-4 by chondrocytes in the surface zone of human osteoarthritic cartilage is regulated by epigenetic DNA de-methylation. Rheumatol Int. 2009;29(5):525–534. doi: 10.1007/s00296-008-0744-z. [DOI] [PubMed] [Google Scholar]
- 15.Bui C, Barter MJ, Scott JL, Xu Y, Galler M, Reynard LN, et al. cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J. 2012;26(7):3000–3011. doi: 10.1096/fj.12-206367. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60(11):3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Andres MC, Imagawa K, Hashimoto K, Gonzalez A, Roach HI, Goldring MB, et al. Loss of methylation in CpG sites in the NF-kappaB enhancer elements of inducible nitric oxide synthase is responsible for gene induction in human articular chondrocytes. Arthritis Rheum. 2013;65(3):732–742. doi: 10.1002/art.37806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynard LN, Bui C, Syddall CM, Loughlin J. CpG methylation regulates allelic expression of GDF5 by modulating binding of SP1 and SP3 repressor proteins to the osteoarthritis susceptibility SNP rs143383. Hum Genet. 2014;133(8):1059–1073. doi: 10.1007/s00439-014-1447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott JL, Gabrielides C, Davidson RK, Swingler TE, Clark IM, Wallis GA, et al. Superoxide dismutase downregulation in osteoarthritis progression and end-stage disease. Ann Rheum Dis. 2010;69(8):1502–1510. doi: 10.1136/ard.2009.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KI, Park YS, Im GI. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J Bone Miner Res. 2013;28(5):1050–1060. doi: 10.1002/jbmr.1843. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Tajes J, Soto-Hermida A, Vazquez-Mosquera ME, Cortes-Pereira E, Mosquera A, Fernandez-Moreno M, et al. Genome-wide DNA methylation analysis of articular chondrocytes reveals a cluster of osteoarthritic patients. Ann Rheum Dis. 2014;73(4):668–677. doi: 10.1136/annrheumdis-2012-202783. [DOI] [PubMed] [Google Scholar]
- 22.Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, et al. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. 2014;66(10):2804–2815. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- 23.Rushton MD, Reynard LN, Barter MJ, Refaie R, Rankin KS, Young DA, et al. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66(9):2450–2460. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Hollander W, Ramos YF, Bos SD, Bomer N, van der Breggen R, Lakenberg N, et al. Knee and hip articular cartilage have distinct epigenomic landscapes: implications for future cartilage regeneration approaches. Ann Rheum Dis. 2014;73(12):2208–2212. doi: 10.1136/annrheumdis-2014-205980. [DOI] [PubMed] [Google Scholar]
- 25.Outerbridge RE. The etiology of chondromalacia patellae. J Bone Joint Surg Br. 1961;43-B:752–757. doi: 10.1302/0301-620X.43B4.752. [DOI] [PubMed] [Google Scholar]
- 26.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Garcia O, Rogers NH, Smith RG, Lotz MK. Palmitate has proapoptotic and proinflammatory effects on articular cartilage and synergizes with interleukin-1. Arthritis Rheumatol. 2014;66(7):1779–1788. doi: 10.1002/art.38399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldring MB, Birkhead JR, Suen LF, Yamin R, Mizuno S, Glowacki J, et al. Interleukin-1 beta-modulated gene expression in immortalized human chondrocytes. J Clin Invest. 1994;94(6):2307–2316. doi: 10.1172/JCI117595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenblatt MB, Ritter SY, Wright J, Tsang K, Hu D, Glimcher LH, et al. NFATc1 and NFATc2 repress spontaneous osteoarthritis. Proc Natl Acad Sci U S A. 2013;110(49):19914–19919. doi: 10.1073/pnas.1320036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamekura S, Kawasaki Y, Hoshi K, Shimoaka T, Chikuda H, Maruyama Z, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54(8):2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 32.Valdes AM, Hart DJ, Jones KA, Surdulescu G, Swarbrick P, Doyle DV, et al. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum. 2004;50(8):2497–2507. doi: 10.1002/art.20443. [DOI] [PubMed] [Google Scholar]
- 33.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 36.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23(3):555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rushton MD, Young DA, Loughlin J, Reynard LN. Differential DNA methylation and expression of inflammatory and zinc transporter genes defines subgroups of osteoarthritic hip patients. Ann Rheum Dis. 2015 doi: 10.1136/annrheumdis-2014-206752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aigner T, Fundel K, Saas J, Gebhard PM, Haag J, Weiss T, et al. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54(11):3533–3544. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18(4):581–592. doi: 10.1016/j.joca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011;23(5):471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotz MK, Carames B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nature reviews Rheumatology. 2011;7(10):579–587. doi: 10.1038/nrrheum.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S, Kim J, Ryu JH, Oh H, Chun CH, Kim BJ, et al. Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat Med. 2010;16(6):687–693. doi: 10.1038/nm.2153. [DOI] [PubMed] [Google Scholar]
- 44.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 45.Brew CJ, Clegg PD, Boot-Handford RP, Andrew JG, Hardingham T. Gene expression in human chondrocytes in late osteoarthritis is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Ann Rheum Dis. 2010;69(1):234–240. doi: 10.1136/ard.2008.097139. [DOI] [PubMed] [Google Scholar]
- 46.Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, Papaioannou VE, et al. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev Cell. 2010;18(1):148–156. doi: 10.1016/j.devcel.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bongers EM, Duijf PH, van Beersum SE, Schoots J, Van Kampen A, Burckhardt A, et al. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74(6):1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu TM, Guo XM, Tan HS, Hui JH, Lim B, Lee EH. Zinc-finger protein 145, acting as an upstream regulator of SOX9, improves the differentiation potential of human mesenchymal stem cells for cartilage regeneration and repair. Arthritis Rheum. 2011;63(9):2711–2720. doi: 10.1002/art.30430. [DOI] [PubMed] [Google Scholar]
- 49.Reynard LN, Bui C, Canty-Laird EG, Young DA, Loughlin J. Expression of the osteoarthritis-associated gene GDF5 is modulated epigenetically by DNA methylation. Hum Mol Genet. 2011;20(17):3450–3460. doi: 10.1093/hmg/ddr253. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalo S. Epigenetic alterations in aging. J Appl Physiol (1985) 2010;109(2):586–597. doi: 10.1152/japplphysiol.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sesselmann S, Soder S, Voigt R, Haag J, Grogan SP, Aigner T. DNA methylation is not responsible for p21WAF1/CIP1 down-regulation in osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2009;17(4):507–512. doi: 10.1016/j.joca.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 53.Taylor SE, Smeriglio P, Dhulipala L, Rath M, Bhutani N. A global increase in 5-hydroxymethylcytosine levels marks osteoarthritic chondrocytes. Arthritis Rheumatol. 2014;66(1):90–100. doi: 10.1002/art.38200. [DOI] [PubMed] [Google Scholar]
- 54.Haseeb A, Makki MS, Haqqi TM. Modulation of ten-eleven translocation 1 (TET1), Isocitrate Dehydrogenase (IDH) expression, alpha-Ketoglutarate (alpha-KG), and DNA hydroxymethylation levels by interleukin-1beta in primary human chondrocytes. J Biol Chem. 2014;289(10):6877–6885. doi: 10.1074/jbc.M113.512269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the design and data analysis used in the study.