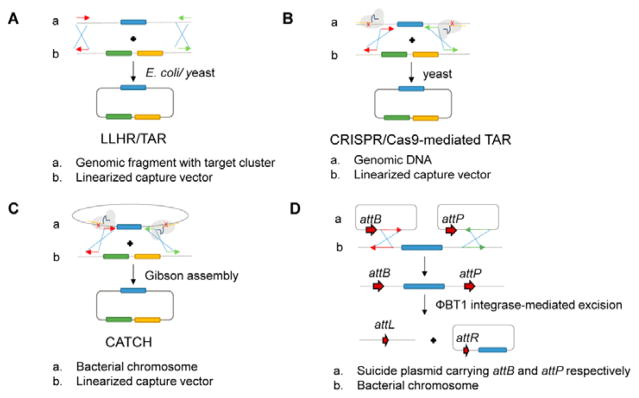
Fig. 5. Methods for direct capture of large DNA fragments.
(A) RecET-mediated linear-linear homologous recombination (LLHR) or transformation-associated recombination (TAR).233, 234 Fragmented/digested genomic DNA with target of interest is assembled with linearized capture vector with homology sequences via HR in either E. coli (LLHR) or yeast (TAR). (B) CRISPR/Cas9-mediated TAR.236 RNA-guided Cas9 is used to cleave chromosomes at designated target sites in vitro. The resultant fragment is assembled with linearized TAR vector in yeast. (C) In CATCH,237 RNA-guided Cas9 is employed to cleave the isolated chromosome at the designated target sites in vitro and the target fragment is captured via Gibson assembly. (D) phiBT1 integrase-mediated site-specific recombination. Suicide plasmids are used to introduce attB and attP recombination flanking the gene locus of interest by HR. Expression of phiBT1 integrase leads to excision of target fragment into a plasmid backbone that is propagated in vivo.55