Abstract
Background To identify differentially expressed miRNAs profiles in bronchoalveolar lavage fluid (BALF) from patients with silicosis and consider the potential contribution of miRNAs to silicosis.
Methods miRNAs expression profiling were performed in the cell fraction of BALF samples obtained from 9 subjects (3 silicosis observation subjects, 3 stage I and stage II silicosis patients, respectively). The differential expression of two selected miRNAs hsa-miR-181c-5p and hsa-miR-29a-3p were confirmed by RT-qPCR. Furthermore, miRNAs Gene Ontology Enrichment categories and target mRNAs were determined based on miRWalk.
Results We found 110 dysregulated miRNAs in silicosis samples, most of which showed a down-regulation trend. Microarray results were confirmed by RT-qPCR. With the observation group samples set as standards, stage I samples showed 123 differentially expressed miRNAs, and stage II 46. 23 miRNAs were dysregulated in both stages. Finally, functional enrichment analysis indicated that these miRNAs played an important role in various biological processes, including ECM-receptor interaction and endocytosis.
Conclusions This is the first time to acquire the BALF-derived microRNAs expression profiling targeting to human silicosis. These results contribute to unravelling miRNAs involved in the pathogenesis of silicosis, and provide new tools of potential use of as biomarkers for diagnosis and/or therapeutic purposes.
Keywords: Silicosis, Bronchoalveolar lavage fluid (BALF), microRNAs (miRNAs), microRNA-29a (miR-29a), microRNA-181c (miR-181c)
Introduction
Silicosis is a systemic disease with main manifestation of pulmonary diffuse fibrosis caused by long-term inhalation and deposition of occupational dust containing silicon dioxide. Silicosis is one of the most common and severe occupational diseases with no effective treatment1, 2, 3). Once developed, even no longer in exposure of silicon dioxide dust, the majority of cases are in progressive development, ultimately resulting in respiratory failure. However, the molecular mechanisms that regulate gene expression in silicosis are incompletely understood, which in a way affects the specific therapeutic methods development4, 5). This highlights the urgent need for application of modern molecular biology techniques to enhance the understanding of silicosis mechanism, which may ultimately help improve the currently disappointing therapy of silicosis patients.
The recent literature suggest that microRNAs (miRNAs) may be important mediators of lung diseases, and more specifically, lung fibrosis6, 7, 8, 9, 10). MiRNAs are 21–25 nucleotides long, small noncoding RNAs, which usually negatively modulate gene expression at the post-transcriptional level by the complete or incomplete complementary binding to target sequences of mRNA11, 12, 13). There is an accumulating body of evidence that miRNAs are involved in a variety of biological processes, such as cell proliferation, differentiation, apoptosis, and tumorigenesis11, 14, 15, 16). Despite the increasing evidence that dysregulation of miRNAs expression has been associated with lung diseases, little is known about profiling of miRNAs in silicosis patients to date.
Our previous study identified a series of miRNAs differentially expressed in rat model of silicosis by using miRNAs expression profiling and found that 39 altered miRNAs in silicosis rats17). In addition, we proved that microRNA-146a and microRNA-181b regulate the secretion of tumor necrosis factor-α and interleukin-1β in silicosis dioxide-induced NR8383 rat macrophages18). Whether these findings could be extrapolated into human silicosis or not has still not been confirmed. Langlois and his colleagues denoted that the miRNAs were different expressed between human and animals19). Besides, different biological samples contains different types of miRNAs. Li et al.7) found that miRNAs in serum samples may play a critical role in the occurrence and development of coal workers’ pneumoconiosis. However, the biomarker in serum can be influenced by many factors, especially diluted by the body fluid, and multiple systems and organs can release the same biomarkers into the serum, not only the lung. Yun et al.20) showed the cytokines and surfactant proteins in bronchoalveolar lavage fluid (BALF) could reflect the inflammation or fibrosis status for coal workers’ pneumoconiosis development better than ones in other body fluids, since it is more sensitive, accurate. More importantly, it can reflect directly the lung status of local inflammation and fibrosis.
So in this study, BALF samples were collected to survey and identify differentially expressed miRNAs from patients at different stages of silicosis. Systematic functional analysis was further performed to explore the potential functions of identified miRNAs correlated with pathogenesis of silicosis.
Subjects and Methods
Study subjects
3 observation subjects and 6 silicosis patients (3 stage I and stage II silicosis patients, respectively) who underwent massive whole lung lavage without clinical contraindications were enrolled in this study. The BALF samples from stage III silicosis patients was absent (or unavailable) because these patients could not hardly survive the massive whole-lung lavage operation. The characteristics of the enrolled subjects are summarized in Table 1. All subjects who had a history of bronchoalveolar lavage, cardiopulmonary dysfunction, active tuberculosis, clotting disorders, severe trachea-bronchial anomalies and beyond 65-year old were excluded. All participants were male. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1983 Helsinki declaration. This work was approved by the Ethical Committee of West China School of Public Health, Sichuan University.
Table 1. The demographic features of the silicosis patients and observation subjects.
| Observation | Stage I | Stage II | P | |
|---|---|---|---|---|
| Age (x±s) | 47.33±3.79 | 45.00±5.00 | 48.33±8.08 | 0.785 |
| Exposure year (x±s) | 11.00±3.61 | 8.33±5.68 | 15.00±7.03 | 0.827 |
| Gender (n, %) | ||||
| Male | 3 (100.0) | 3 (100.0) | 3 (100.0) | 1.000 |
| Smoking (n,%) | 0.638 | |||
| No | 2 (66.7) | 1 (33.3) | 1 (33.3) | |
| Yes | 1 (33.3) | 2 (66.7) | 2 (66.7) | |
| Drinking (n,%) | 1.000 | |||
| No | 2 (66.7) | 2 (66.7) | 2 (66.7) | |
| Yes | 1 (33.3) | 1 (33.3) | 1 (33.3) | |
| Pulmonary function (%) | ||||
| VC | 84.03±14.06 | 80.20±7.43 | 85.20±13.06 | 0.868 |
| FVC | 86.33±14.22 | 82.10±7.76 | 85.43±130.3 | 0.903 |
| FEV1 | 96.80±14.26 | 88.87±16.46 | 87.97±13.85 | 0.738 |
| DLco | 90.70±21.12 | 84.70±21.69 | 83.50±7.77 | 0.875 |
| RV | 103.07±17.30 | 146.10±32.50 | 132.47±19.96 | 0.164 |
| RV/TLC | 36.57±1.38 | 46.12±4.39 | 45.33±5.54 | 0.055 |
Severity of silicosis definition
The diagnosis of silicosis was established by a combination of typical clinical and radiological findings on high quality X-ray based on the diagnostic criteria of pneumoconiosis (GBZ 70-2009) in China, which were based on the 2000 ILO classification. The criteria describe the disease in stages 0, I, II, and III, referring to none, mild, moderate, and severe, respectively. Stages 0, I, and II correspond to 0, 1–2, and 3 by the ILO classification; stage III corresponds to pneumoconiosis with large opacities (categories A, B and C by the ILO classification). In the circumstances, Stage 0 workers could be chosen as the control group since Stage 0 are called as observation subjects whose radiological findings on X-ray cannot be identified as stages I and significant clinic symptoms cannot be detected but X-ray imaging changes need follow-up.
Clinical specimens
BALF samples were obtained by instillation and recovery of 0.9% sterile saline in the bronchopulmonary segment21). About 3,000 mL recovered fluid was condensed immediately by centrifuged at 600×g at RT for 15 min (Shuke, Sichuan, China), and the most supernatants was discarded. Cells were isolated by density gradient centrifugation (Thermo Fisher Scientific, Rockford, IL, USA). The pellets were filtrated by sterilized double-layer gauze to remove mucus and the filtrate were centrifuged at 1,500×g at 4°C for 15 min, then the pellets from these samples was washed three times by Hank’s (Sigma, St. Louis, MO, USA). Cells layer were pipetted into eppendorf tubes and centrifuged 10 min at 1,500×g at 4°C. Finally, the cells fraction was resuspended in 1 mL RNA Store (Tiangen, Beijing, China). According to the above treatment, about 90 percent cells has already been confirmed as macrophages.22)
RNA isolation and RNA quality control
Total RNA containing small RNA was extracted from bronchoalveolar cells fraction using a miRVana miRNA isolation kit (Applied Biosystems, Foster, CA, USA). RNA quality and quantity were measured by NanoDrop 1,000 spectrophotometer (Thermo Fisher Scientific, Rockford, IL, USA) and RNA Integrity was determined by gel electrophoresis. All RNA samples showed high quality without RNA degradation or DNA contamination.
MiRNAs microarray analysis
After RNA isolation from the samples, the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) was used according to the manufacturer’s guideline for miRNA labeling. After terminating the labeling procedure, the Hy3TM-labeled samples were hybridized on the miRCURYTM LNA Array (v.18.0) (Exiqon, Vedbaek, Denmark), which contains 3,100 capture probes, covering all human, mouse and rat microRNAs annotated in miRBase 18.0, as well as all viral microRNAs related to these species according to array manual. Following the washing steps the slides were scanned using the Axon GenePix 4000B microarray scanner. Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs whose intensities>=30 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the Median normalization. After normalization, significant differentially expressed miRNAs were identified through Volcano Plot filtering. Hierarchical clustering was performed using MEV software (v4.8, TIGR).
Quantitative real-time PCR (RT-qPCR)
To verify the array results, we collected another 3 observation subjects and 6 patients with stage I and stage II silicosis using RT-qPCR to detect the expression of hsa-miR-181c-5p and hsa-miR-29a-3p. Total RNA (150 ng), 1 μM RT primer (Table 2), 10×RT buffer, 2.5 mM each of dATP, dGTP, dCTP and dTTP (HyTest Ltd., Turku, Finland), 200 U/μl reverse transcriptase (Epicentre, Palmerston North, New Zealand) and 40 U/μl RNase Inhibitor (Epicentre, Palmerston North, New Zealand) were subjected to reverse transcriptase reactions. PCR master mix (Qiagen, Hilden, Germany) and 0.6 μM of each primer (Table 1) were used for RT-qPCR. Each experiment was performed in triplicate.
Table 2. Oligonucleotides used in the study.
| Primer set name | Reverse transcriptase reaction primer | Real-time quantitative PCR primer |
|---|---|---|
| U6 | 5’CGCTTCACGAATTTGCGTGTCAT3’ | F:5’GCTTCGGCAGCACATATACTAAAAT3’ |
| R:5’CGCTTCACGAATTTGCGTGTCAT3’ | ||
| hsa-miR-181c-5p | 5’GTCGTATCCAGTGCGTGTCGTGGAGTC | GSP:5’ GGGAACATTCAACCTGTCG3’ |
| GGCAATTGCACTGGATACGACACTCAC3’ | R:5’GTGCGTGTCGTGGAGTCG3’ | |
| hsa-miR-29a-3p | 5’ GTCGTATCCAGTGCGTGTCGTGGAGTC | GSP:5’ GGGGTAGCACCATCTGAAAT3’ |
| GGCAATTGCACTGGATACGACTAACCG3’ | R:5’GTGCGTGTCGTGGAGTCG3’ |
MiRNAs target gene prediction and functional analysis
We selected a series of abundantly and differentially expressed miRNAs to query for their Gene Ontology Enrichment categories using the Visualization and Integrated Discovery (DAVID) V6.7 bioinformatics resources (http://david.abcc.ncifcrf.gov/)23). The target mRNAs of these miRNAs were determined based on miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), which is a comprehensive database that provides information on predicted as well as validated binding sites on their target genes miRNA from Human, Mouse and Rat. MiRNA-targets interactions information were produced by 8 established miRNA prediction programs on 3’ UTRs of all known genes of Human, Mouse and Rat i.e. RNA22, miRanda, miRDB, TargetScan, RNAhybrid, PITA, PICTAR, and Diana-microT24).
Statistical analysis
The R software from the “The Comprehensive R Archive Network” was used to perform statistical processing. Due to the small sample size, we adopted the random variance model t-test to filter the differentially expressed miRNAs between silicosis patients and observation subjects. The differentially expressed miRNA were considered statistically significant if they were up or down-regulated at least 2-fold. We distinguished the different miRNAs using significant analysis and false discovery rate (FDR) analysis to correct the P value. The significant level was 0.05 (two-tailed).
Results
Differentially expressed miRNAs profiles
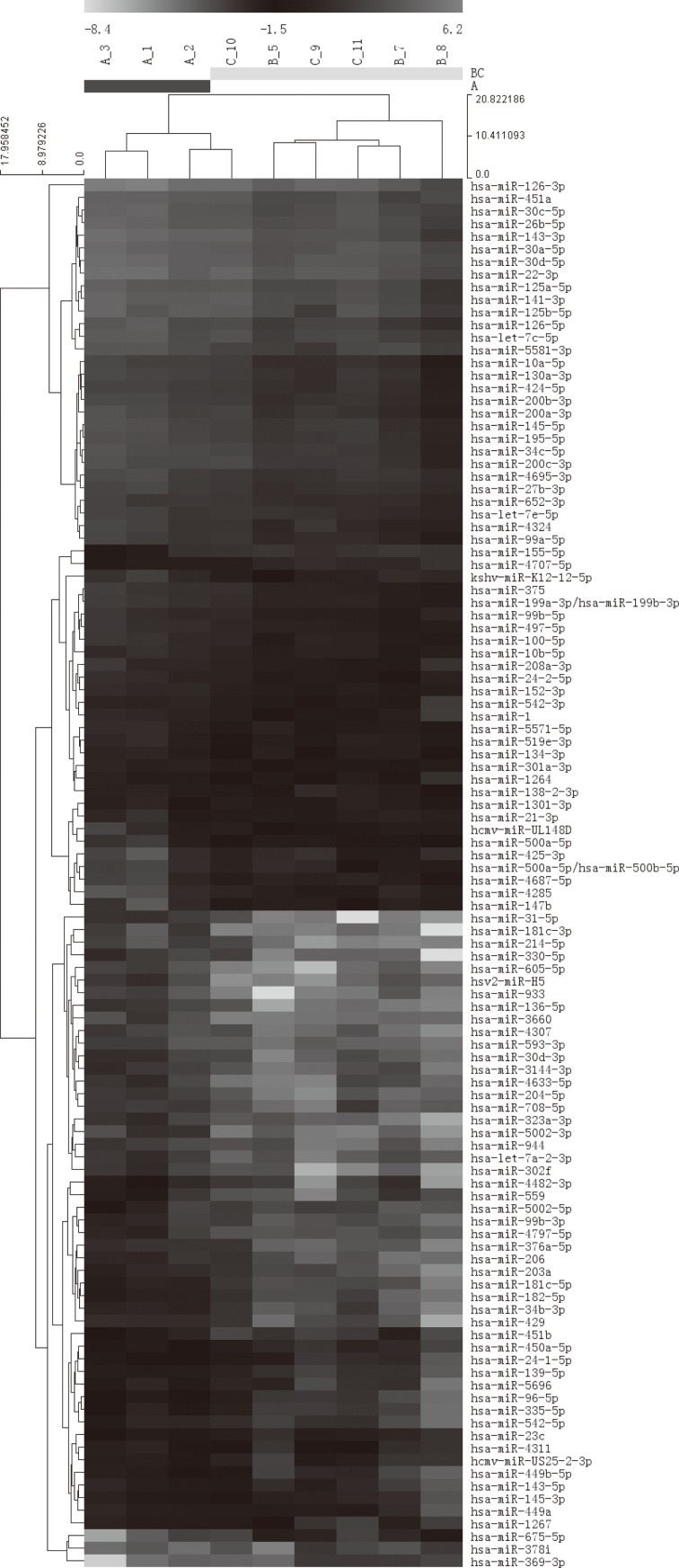
By applying a high-sensitive, specific and high-throughput miRCURYTM LNA microarray, the bronchoalveolar fluid cytology-derived miRNA profiles of silicosis patients were mapping. Compared with observation subjects, silicosis groups including two stages showed significantly distinct miRNAs distribution. We found that out of ~1,900 annotated miRNAs in miRBase 18.0 as well as all viral miRNAs (~150) related to human miRNAs that were confidently detected, 110 were dysregulated in silicosis samples. Finally, hierarchical clustering was analyzed to show distinguishable miRNA expression profiling among samples (Fig. 1).
Fig. 1.
Hierarchical clustering for differentially expressed miRNA in silicosis group and observation subjects pass Volcano Plot (Fold Change>=2, each row represents a miRNA and each column represents a sample. 9 samples were clustered according to the expression profile of 110 differentially expressed miRNAs. The miRNA clustering tree is shown on the left, and the sample clustering tree appears at the top. Red color represents a high relative expression level; green color represents a low relative expression level. )
Validation of microarray results using RT-qPCR
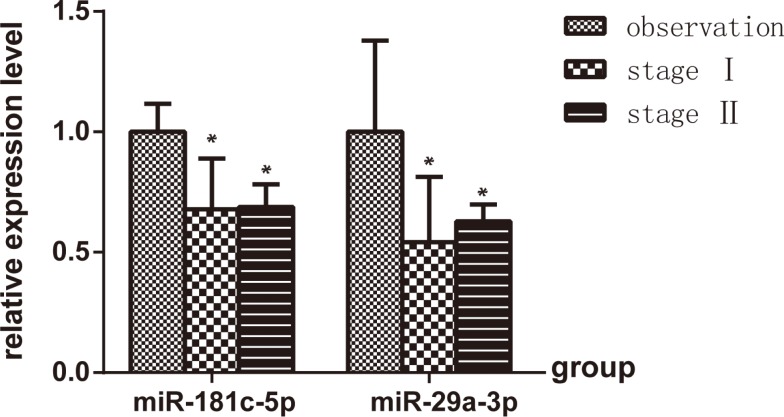
In order to validate the microarray results, quantitative real-time PCR analysis of bronchoalveolar cell fraction-derived miRNA expression from study subjects was completed for hsa-miR-181c and has-miR-29a. Compared with observation subjects, the expression of hsa-miR-181c and hsa-miR-29a was significantly lower in stage I and Stage II silicosis patients (P<0.05), but there was no significantly difference between stage I and Stage II silicosis patients (P>0.05) (Fig. 2), furthermore, the trend was consistent with the microarray results.
Fig. 2.
Distribution of the has-miR-181c and has-miR-29a across stage I (n=3) and stage II (n=3) based on the observation samples (n=3). *P<0.05, vs. observation.
Differentially expressed miRNAs in two different stages
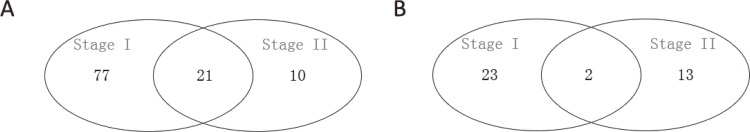
The observation group samples setting as standards, differentially expressed miRNAs among the two stages were determined and characterized. Stage II samples showed 46 (15 up-regulated and 31 down-regulated) and stage I samples 123 (25 up-regulated and 98 down-regulated) changed miRNAs. We listed 23 miRNAs which are abundantly expressed in both two groups (Table 3), for example, hsa-miR-31-5p was significantly down-regulated and hsa-miR-1301-3p was significantly up-regulated. Interestingly, we found that significantly down-regulated miRNAs occurred more frequently than significantly up-regulated miRNAs. We also found 23 miRNAs were dysregulated in both stages (up-regulated: 2miRNAs; down-regulated: 21 miRNAs) (Fig. 3). We also conducted pairwise comparisons to further characterize the differentially expressed miRNAs across two different stages (Table 4). Based on the stage I samples, stage II samples showed 18 differentially expressed miRNAs (16 up-regulated and 2 down-regulated).
Table 3. The aberrantly expressed miRNA profiles based on the observation samples.
| miRNA* | Stage I/observation | Stage II/ observation |
|---|---|---|
| hsa-miR-500a-5p | 4.82 | 4.50 |
| hsa-miR-1301-3p | 4.17 | 5.98 |
| hsa-miR-24-1-5p | 0.20 | 0.23 |
| hsa-miR-3144-3p | 0.15 | 0.19 |
| hsa-miR-152-3p | 0.14 | 0.24 |
| hsa-miR-5571-5p | 0.14 | 0.16 |
| hsa-miR-450a-5p | 0.13 | 0.21 |
| hsa-miR-204-5p | 0.11 | 0.17 |
| hsa-miR-497-5p | 0.09 | 0.21 |
| hsa-miR-24-2-5p | 0.08 | 0.23 |
| hsa-miR-195-5p | 0.08 | 0.24 |
| hsa-miR-323a-3p | 0.07 | 0.15 |
| hsa-miR-34c-5p | 0.07 | 0.24 |
| hsa-miR-199a-3p | 0.07 | 0.22 |
| hsa-miR-203a | 0.06 | 0.25 |
| hsa-miR-181c-5p | 0.05 | 0.17 |
| hsa-miR-96-5p | 0.05 | 0.24 |
| hsa-miR-1 | 0.03 | 0.21 |
| hsa-miR-182-5p | 0.03 | 0.20 |
| hsa-miR-34b-3p | 0.02 | 0.25 |
| hsa-miR-136-5p | 0.02 | 0.15 |
| hsa-miR-31-5p | 0.01 | 0.06 |
| hsa-miR-1267 | 0.15 | 0.07 |
*The listed miRNAs are abundantly expressed in both two groups.
Fig. 3.
Distribution of differentially expressed miRNAs between two different stages of silicosis (A. Down-regulated miRNAs B. Up-regulated miRNAs)
Table 4. miRNAs that were differentially expressed across the two different stages of silicosis.
| miRNA | Stage II/ Stage I |
|---|---|
| hsa-miR-542-5p | 4.69 |
| hsa-miR-1255b-2-3p | 6.36 |
| hsv1-miR-H7-3p | 4.06 |
| hsa-miR-126-5p | 4.53 |
| hsa-miR-10b-5p | 6.59 |
| hsa-miR-5093 | 28.25 |
| hsa-miR-4268 | 6.19 |
| hsa-miR-660-3p | 4.89 |
| hsa-miR-126-3p | 4.17 |
| hsa-let-7f-5p | 4.11 |
| hsa-miR-429 | 7.73 |
| hsa-miR-100-5p | 4.63 |
| hsa-miR-4311 | 5.17 |
| hsa-miR-199b-5p | 4.99 |
| hsa-miR-3679-3p | 0.18 |
| ebv-miR-BHRF1-1 | 0.20 |
| hsa-miR-34b-3p | 6.45 |
| hsa-miR-449b-5p | 8.17 |
Functional enrichment analysis to predicting potential target mRNAs
We selected abundantly expressed miRNAs to further query for functional enrichment analysis according to their experimentally validated targets mRNAs. The results showed that these miRNAs possibly contributed to multiple regulation networks and played an important role in various essential biological processes, especially involved in endocytosis and focal adhesion including ECM-receptor interaction (Table 5).
Table 5. Enrichment pathway analysis identified target mRNAs of abundantly and differentially expressed miRNAs.
| Pathway | Count | P value | Target mRNAs |
|---|---|---|---|
| Focal adhesion | 14 | 1.2E-2 | PARVG, MET, PRKCG, PPP1CC, CHAD, VCL, ITGA6, PAK4, GSK3B, PDGFRA, PIK3R5, MYLK, FN1, PARVA |
| Neurotrophin signaling pathway | 12 | 2.0E-3 | IRAK3, BDNF, YWHAZ, RPS6KA4, RPS6KA1, MAP3K1, GSK3B, RIPK2, PIK3R5, NGFR, CALM2, TP73 |
| Regulation of actin cytoskeleton | 12 | 8.6E-2 | CHRM5, ITGA6, PAK4, MRAS, PDGFRA, CYFIP2, PIK3R5, PPP1CC, MYLK, VCL, TTLL3, FN1 |
| Endocytosis | 11 | 7.2E-2 | FAM125B, CBLB, VPS45, MET, PSD3, PDGFRA, VPS4A, VPS37B, PSD2, SMURF1, CSF1R |
| Vascular smooth muscle contraction | 8 | 6.9E-2 | ADCY1, PTGIR, PRKCG, CALCRL, PPP1CC, PRKCE, CALM2, MYLK |
| Long-term potentiation | 7 | 2.2E-2 | GRM5, ADCY1, RPS6KA1, PPP3R2, PRKCG, PPP1CC, CALM2 |
| Melanogenesis | 7 | 9.9E-2 | DVL3, ADCY1, WNT7B, GSK3B, CREB3L1, PRKCG, CALM2 |
| Phosphatidylinositol signaling system | 6 | 8.9E-2 | DGKE, PIK3R5, DGKH, PRKCG, INPP5D, CALM2 |
Discussion
To date, several research groups have performed functional studies that have confirmed the important role of miRNAs deregulation in lung diseases8, 25, 26, 27, 28, 29). MiRNAs have been described as ideal biomarkers due to their stability and the feasibility to quantify miRNAs in a standardized manner30, 31). To further clarify the role of miRNAs in pathogenesis of silicosis, we performed genome-wide miRNAs expression profiling in BALF cell fraction of 3 silicosis observation subjects and 6 silicosis patients belonging to stage I (n=3) and stage II (n=3). A total of 110 dysregulated microRNAs were found to be significantly differentially expressed in bronchoalveolar fluid cytology-derived samples from silicosis patients. With the observation group samples set as standards, stage I samples showed 123 and stage II samples showed 46 differentially expressed miRNAs. Though the sampling size was small, the result of microchip could give us a specific hint to find potential biomarkers to human silicosis. These data show that expression of miRNAs is altered in silicosis and that different stages of silicosis are associated with distinct changes in miRNAs expression.
Not surprisingly, in concordance with previous animal model data17), we also found significant down-regulated miRNAs was a more common trend in silicosis patients. Together, these results suggest that the phenomena of down-regulation might partly contribute to inflammation and fibrosis of human silicosis. Moreover, the stage I and stage II shared 23 common miRNAs profiling. In order to further analyze the altered miRNAs, differentially expressed miRNAs were also subjected to pairwise comparison between the two different stages. Based on the stage I samples, stage II samples showed 18 differentially expressed miRNAs, which may be the consequence of different degree of inflammation and fibrosis between two stages. The dynamic expression profiles across two different stages of silicosis also indicate a potential contribution of small RNAs in pathogenesis of silicosis, especially for the progression through the different stages of the disease. But, compared with previous data17), the miRNA profile pattern of silicosis patients was differences, and they only expressed few same miRNA family, such as miR-181 and miR-29 as silicosis rats. These may result from the different species and different disease severity. Silicosis rat models all have more severe fibrosis than silicosis patients, which may contribute to the results.
More recently, abnormalities of expression of multiple miRNAs in lung diseases have been reported. Akin to a landmark study, some studies have demonstrated that miR-29 is a major regulator of genes associated with pulmonary fibrosis6, 32, 33). According to our study, we also found significant down-regulation of miR-29 in silicosis patients, suggesting that the down-regulation of miR-29 expression may be indispensable to the pathophysiology of silicosis. Our previous study showed that miR-181b regulate the secretion of tumor necrosis factor-α and interleukin-1β in silicosis dioxide-induced NR8383 rat macrophages18). Previous study also illustrated that miR-181c was involved in a tumor-suppression pathway34). In this study, we also found miR-181c down-regulated in silicosis group using RT-qPCR. Since different miR-181 family members such as miR-181b and miR-181c share a common seed and differ in other miRNA regions, miR-181 family played a vital role in silicosis35, 36). Besides, we also found miR-24, miR-204, miR-34, miR-96 and miR-1 were dysregulated in silicosis group, which have been reported to be involved in other lung diseases37, 38, 39, 40, 41). Further studies, especially for experimental validation, should reveal the potential molecular mechanisms of these miRNAs how to regulate the mRNAs expression to promote or inhibit the progress of silicosis.
Furthermore, Gene Ontology (GO) analysis was applied to gain significant insight into the molecular function and biological process of these target genes. The results suggest that these aberrantly expressed miRNAs have participated in various biological processes. The adhesive interactions of cells with ECM molecules have wide-ranging effects on cellular differentiation and tissue structure in vivo42). At the molecular level, these adhesion-dependent signals are mediated by clustering of integrin adhesion receptors and organization of the actin cytoskeleton into specialized morphological structures, including focal adhesions and actin cytoskeleton. Numerous in vivo and in vitro studies have indicated that alveolar macrophage (AM) undergo an “activation” process in response to lung injury, in which they swallow dust and produce extracellular matrix4, 43). The essential biological roles, especially involved in endocytosis and focal adhesion including ECM-receptor interaction, implicate the potential crucial roles of miRNAs in silicosis.
To our knowledge, this is the first study to analyze the differentially expression of bronchoalveolar fluid cytology-derived miRNAs profiles from silicosis patients. Compared with serum samples, the BALF samples have priority to lung diseases research on account of its sensitivity and accuracy reflecting pulmonary micro-environment. These differentially expressed microRNAs were probable candidates involved in the pathogenesis of silicosis, which would be used in the diagnosis or treatment of the disease in the future. However, our study had several limitations. Firstly, it was a single-center study involving a small sample size. Secondly, lung lavage fluid cannot be obtained from healthy individual and stage III silicosis patients which may underestimate or overestimate the differences. Finally, the concomitant medication was not registered or controlled for. Therefore, large-scale multicenter studies including additional clinical information are required to further verify associations between miRNAs expression and clinical features and outcome of patients with silicosis. In any case, it is important to highlight that our results clearly identified discriminating miRNA expression patterns for silicosis. In addition, the results provide an elementary background to dig into the role of miRNAs in the complex mechanisms of silicosis pathogenesis and its potential implications in strategies for disease control. It is expected that manipulating altered miRNAs expression using synthetic miRNA or antagomirs might become a viable therapeutic approach as part of future “personalized” or precision treatments of silicosis.
In summary, our study initially found differentially expressed miRNAs profiles in silicosis patients and dynamic expression patterns across the different stages of silicosis. Moreover, aberrantly expressed miRNAs have multiple roles in various biological processes, including ECM-receptor interaction and endocytosis. It may be concluded that the aberrant expression of these important miRNAs may contribute to the occurrence and development of silicosis. Their potential contribution and mechanistic details need to be further studied.
Acknowledgments
This study was supported by the National Nature Science Foundation of China (No: 81102107, 81202181).
References
- 1.Cohen RA, Patel A, Green FH. Lung disease caused by exposure to coal mine and silica dust; 2008. © Thieme Medical Publishers. pp. 651–61. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg MI, Waksman J, Curtis J (2007) Silicosis: a review. Dis Mon 53, 394–416. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CR, Kelley TR (2010) A brief review of silicosis in the United States. Environ Health Insights 4, 21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rimal B, Greenberg AK, Rom WN (2005) Basic pathogenetic mechanisms in silicosis: current understanding. Curr Opin Pulm Med 11, 169–73. [DOI] [PubMed] [Google Scholar]
- 5.Yucesoy B, Vallyathan V, Landsittel DP, Simeonova P, Luster MI (2002) Cytokine polymorphisms in silicosis and other pneumoconioses. Mol Cell Biochem 234–235, 219–24. [PubMed] [Google Scholar]
- 6.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lü J (2011) miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 45, 287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Ji X, Yang S, Hou Z, Luo C, Fan J, Ni C, Chen F (2013) Genome-wide analysis of aberrantly expressed circulating miRNAs in patients with coal workers’ pneumoconiosis. Mol Biol Rep 40, 3739–47. [DOI] [PubMed] [Google Scholar]
- 8.Tomankova T, Petrek M, Kriegova E (2010) Involvement of microRNAs in physiological and pathological processes in the lung. Respir Res 11, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J (2013) miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One 8, e60317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke Y, Zhao W, Xiong J, Cao R (2013) Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS Lett 587, 3153–7. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. cell 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 13.He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5, 522–31. [DOI] [PubMed] [Google Scholar]
- 14.Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11, 597–610. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM (2006) MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 66, 7390–4. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–66. [DOI] [PubMed] [Google Scholar]
- 17.Faxuan W, Qin Z, Dinglun Z, Tao Z, Xiaohui R, Liqiang Z, Yajia L (2012) Altered microRNAs expression profiling in experimental silicosis rats. J Toxicol Sci 37, 1207–15. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wang F, Lan Y, Zhou D, Ren X, Zhao L, Zhang Q (2015) Roles of microRNA-146a and microRNA-181b in regulating the secretion of tumor necrosis factor-α and interleukin-1β in silicon dioxide-induced NR8383 rat macrophages. Mol Med Rep 12, 5587–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langlois RA, Albrecht RA, Kimble B, Sutton T, Shapiro JS, Finch C, Angel M, Chua MA, Gonzalez-Reiche AS, Xu K, Perez D, García-Sastre A, tenOever BR (2013) MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat Biotechnol 31, 844–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Wang H, Xing J, Liu Y, Cui X, Guo J, Chen W (2014) Expression levels of surfactant-associated proteins and inflammation cytokines in serum and bronchoalveolar lavage fluid among coal miners: a case-control study. J Occup Environ Med 56, 484–8. [DOI] [PubMed] [Google Scholar]
- 21.Molina-Pinelo S, Suárez R, Pastor MD, Nogal A, Márquez-Martín E, Martín-Juan J, Carnero A, Paz-Ares L (2012) Association between the miRNA signatures in plasma and bronchoalveolar fluid in respiratory pathologies. Dis Markers 32, 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Zhao J, Fu B, Zhang J (1994) Analysis of bronchoalveolar lavage fluid in workers with pneumoconiosis. Zhonghua lao dong wei sheng zhi ye bing za zhi=Zhonghua laodong weisheng zhiyebing zazhi=Chinese journal of industrial hygiene and occupational diseases 12, 209–11. [Google Scholar]
- 23.Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 24.Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44, 839–47. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, Liu X, Nelson A, Nakanishi M, Kanaji N, Wang X, Kim M, Li Y, Sun J, Michalski J, Patil A, Basma H, Holz O, Magnussen H, Rennard SI (2010) Reduced miR-146a increases prostaglandin E2 in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med 182, 1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Pottelberge GR, Mestdagh P, Bracke KR, Thas O, van Durme YM, Joos GF, Vandesompele J, Brusselle GG (2011) MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 183, 898–906. [DOI] [PubMed] [Google Scholar]
- 27.Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA (2009) Divergent intracellular pathways regulate interleukin-1β-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett 583, 3349–55. [DOI] [PubMed] [Google Scholar]
- 28.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64, 3753–6. [DOI] [PubMed] [Google Scholar]
- 29.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA (2008) Rapid changes in microRNA-146a expression negatively regulate the IL-1β-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 180, 5689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brock M, Rechsteiner T, Kohler M, Franzen D, Huber LC (2015) Kinetics of microRNA Expression in Bronchoalveolar Lavage Fluid Samples. Lung 193, 381–5. [DOI] [PubMed] [Google Scholar]
- 32.Pandit KV, Milosevic J, Kaminski N (2011) MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 157, 191–9. [DOI] [PubMed] [Google Scholar]
- 33.Xiao J, Meng XM, Huang XR, Chung ACK, Feng YL, Hui DSC, Yu CM, Sung JJY, Lan HY (2012) miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther 20, 1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Papagiannakopoulos T, Puskar K, Qi S, Santiago F, Clay W, Lao K, Lee Y, Nelson SF, Kornblum HI, Doyle F, Petzold L, Shraiman B, Kosik KS (2007) Detection of a microRNA signal in an in vivo expression set of mRNAs. PLoS One 2, e804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neel JC, Lebrun JJ (2013) Activin and TGFβ regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell Signal 25, 1556–66. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Wan X, Gu Z, Zhang H, Yang X, He L, Miao R, Zhong Y, Zhao H (2014) Evolution of the mir-181 microRNA family. Comput Biol Med 52, 82–7. [DOI] [PubMed] [Google Scholar]
- 37.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–98. [DOI] [PubMed] [Google Scholar]
- 38.Courboulin A, Paulin R, Giguère NJ, Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher S, Côté J, Simard MJ, Bonnet S (2011) Role for miR-204 in human pulmonary arterial hypertension. J Exp Med 208, 535–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, Tan X, Lu N, Sun Y, Sun J, Sun N, He J (2011) DNA hypermethylation of microRNA-34b/c has prognostic value for stage I non-small cell lung cancer. Cancer Biol Ther 11, 490–6. [DOI] [PubMed] [Google Scholar]
- 40.Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, Yoshino H, Kawahara K, Nishiyama K, Seki N, Nakagawa M (2011) MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci 102, 522–9. [DOI] [PubMed] [Google Scholar]
- 41.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K (2008) Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem 283, 33394–405. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Hynes RO. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–87. [DOI] [PubMed] [Google Scholar]
- 43.Davis GS. (1986) Pathogenesis of silicosis: current concepts and hypotheses. Lung 164, 139–54. [DOI] [PubMed] [Google Scholar]