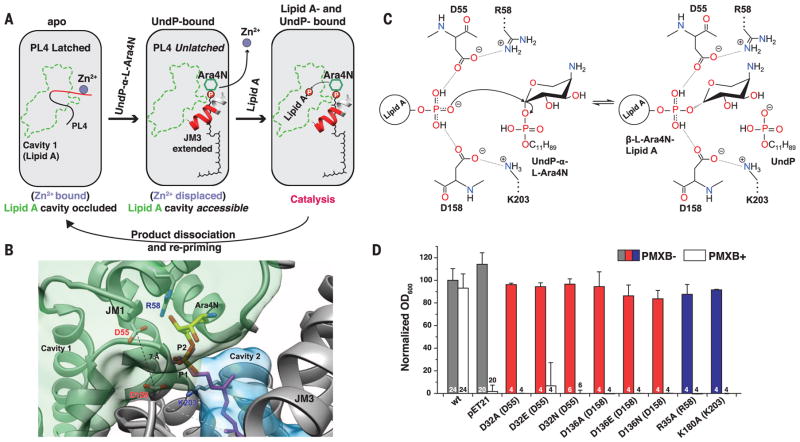
Fig. 4. Putative catalytic mechanism of ArnT.
(A) Schematic representation of substrate-binding–induced conformational changes and catalytic cycle of ArnTCm. The boundaries of cavity 1 are shown in green, and the loop (PL4) that rearranges upon UndP binding is in red. (B) Structural perspective of the active site of ArnTCm. A putative position for the aminoarabinose sugar determined by docking is shown. The conserved D55 and D158 are located at the interface between the binding site for UndP-L-Ara4N and cavity 1, and have a Cγ-Cγ distance of 7.1 Å. P1 (magenta) is the phosphate of experimentally determined UndP, whereas P2 (heteroatom) is the phosphate from the modeled L-Ara4N-phosphate. (C) Putative catalytic mechanism in which two of the oxygen atoms of the acceptor phosphate are coordinated by D55 and D158, which leaves the third with a net negative charge, primed for a direct nucleophilic attack on the arabinose ring. This mechanism is consistent with inversion of the glycosidic bond, as reported for L-Ara4N attachment to lipid A (23). A precedent for a phosphate acting as a nucleophile exists (24). R58 and K203 appear to act similarly as the catalytic Mg2+, in the case of PglB, that localizes the charge of an aspartate and a glutamate, which in turn coordinate the nucleophile acceptor amide (19, 25). (D) Function of putative catalytic aspartates D32 and D136 and positively charged residues R35 and K180 in ArnTSe tested utilizing the PMXB growth assay (13). Data presented are means + SD. N is shown for each data column.