Abstract
KIAA0319 at the DYX2 locus is one of the most extensively studied candidate genes for developmental dyslexia (DD) owing to its important role in neuronal migration. Previous research on associations between KIAA0319 genetic variations and DD has yielded inconsistent results. It is important to establish a more precise estimate of the DD risk associated with these genetic variations. We carried out a meta-analysis of association studies involving KIAA0319 polymorphisms and DD risk. The results of pooled analysis indicated that none of the six investigated markers in or near the KIAA0319 gene are associated with DD. However, a stratified analysis by the study population revealed opposite associations involving KIAA0319 rs4504469 in European and Asian subgroups. The stratified analysis also showed that the KIAA0319 rs9461045 minor allele (T allele) has a protective effect in Asians. This meta-analysis has allowed us to establish the effects of specific KIAA0319 polymorphisms on DD risk with greater precision, as they vary across populations; analyzing one single nucleotide polymorphism at a time could not fully explain the genetic association for DD.
Developmental dyslexia (DD) is one of the most common learning disorders affecting 5–10% of school-aged children worldwide1. It is characterized by a specific impairment in reading in the context of adequate intelligence and educational opportunity1. Longitudinal evidence indicates that individuals who are diagnosed with DD exhibit long-term, persistent deficits in several cognitive skills and functions, such as phonology, orthography, attention, memory and perception2,3,4,5,6. These deficits hinder access to knowledge and have an adverse effect on the educational achievement and socioeconomic status of sufferers across the life span7,8. Although the pathology of DD remains elusive, it is generally considered to be neurobiological in origin6,9, and is highly heritable10,11. Initial genetic linkage studies have identified at least nine DD susceptibility loci referred to as DYX1 to DYX9; the most consistently replicated of these is DYX2, which lies at chromosome 6p22.212,13,14,15. This locus has been the subject of numerous studies from which KIAA0319 has emerged as one of the two strongest candidates (the other is DCDC2) for DD16,17,18,19,20,21,22.
During the past decade some noteworthy genetic variants in KIAA0319 have been identified in independent DD samples. An early study by Francks et al. identified an underlying quantitative trait locus at chromosome 6p22.2, a 77 kb region spanning the entire TTRAP and the first four exons of KIAA0319, which was found to be related to DD in two independent UK samples and one US sample; the authors suggested that a three-marker risk haplotype (rs4504469-rs2038137-rs2143340) in this region was associated with DD16, thereby identifying KIAA0319 as a DD-associated gene. Following this study, rs4504469, rs6935076 and their haplotype were observed to be associated with DD in a UK sample23. Subsequently, Harold et al. not only confirmed that rs4504469 and rs6935076 influenced DD risk, but also identified another promising single nucleotide polymorphism (SNP) candidate for association with DD, rs761100, in a larger UK sample24. However, the significant associations of rs4504469 and rs6935076 with DD risk were not confirmed in a US sample studied by Brkanac et al.25. The positive effect of rs4504469 and rs761100 also did not occur in Toronto samples investigated by Couto et al.19, in which they found that the alternate allele of rs6935076 was in fact associated with elevated DD risk, which contrasts with findings in the two previously reported UK samples23,24. The opposite effects were also found for the rs4504469 T allele on DD risk. Three years ago, we combined results from five independent studies of European samples26 and showed that the rs4504469 T allele had a protective effect with respect to DD. However, subsequent researches in Asian samples27,28 showed that the T allele conferred an increased risk of DD.
Nonetheless, KIAA0319 seems to be an important risk factor for DD, and this conclusion is supported by functional analysis. KIAA0319 encodes for an integral membrane protein containing a large extracellular domain with four polycystic kidney disease domains, a single transmembrane domain and a small intracellular C-terminus26,29. Evidence from the cellular level indicates that the KIAA0319 protein may be involved in signaling as well as cell-cell interactions30,31,32. In addition in animal model studies, an embryonic Kiaa0319 knockdown resulted in disruption to the migration of neocortical neurons leading to the formation of heterotopia in white matter in a subset of animals which also displayed an array of behavioral deficits, including impairments in rapid auditory processing, simple spatial learning33 and phoneme processing34, similar to those seen in dyslexics. Disrupted neural migration is thought to be an important feature of DD35. This evidence suggests that the level of KIAA0319 expression plays an important role in the development of DD.
Several SNPs are involved in regulation of KIAA0319 expression. The risk haplotype rs4504469-rs2038137-rs2143340 identified by Francks et al.16 has been linked to lower levels of KIAA0319 transcripts17,36, and rs9461045 has been associated with reduced gene expression37 in experimental studies, suggesting that expression of this gene may be affected by the component markers, or other polymorphisms in linkage disequilibrium (LD) with these markers (such as rs3212236 in LD with rs9461045).
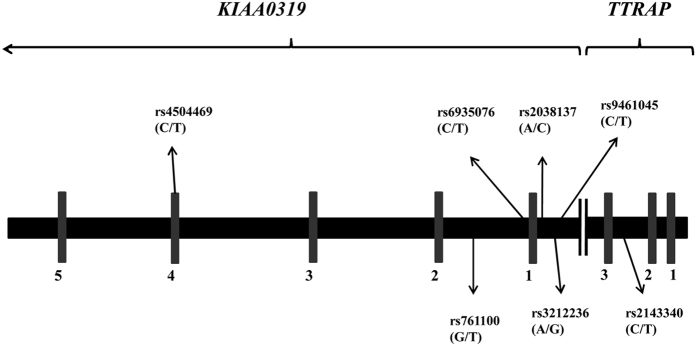
The inconsistent results of previous association studies may be due to the limited statistical power of single studies, which means that the modest effects of these variants on DD risk may go undetected owing to the small sample sizes. Meta-analyses are an important tool in the field of association studies, especially when there are lots of published studies and results are inconsistent across studies. We therefore conducted a meta-analysis integrating the results from case-control and transmission/disequilibrium test (TDT) studies in order to derive a more precise estimate of the association between six markers in or near the KIAA0319 gene (Fig. 1) and DD risk. The criteria for the choice of SNPs were that 1) they had been subject of numerous, inconsistent reports, e.g. rs4504469, rs761100 and rs6935076, or were likely to be related to regulation of KIAA0319 expression, e.g. rs2143340, rs9461045 and rs3212236; and 2) they had been the subject of at least four relevant association studies. We did not include rs2038137 as only three association studies had been published. Because of the inconsistent findings in relation to KIAA0319 markers and DD risk in different populations we conducted a stratified analysis for all included polymorphisms, with stratification based on the study population.
Figure 1. Gene structure and location of six single nucleotide polymorphisms (SNPs) in or near the KIAA0319 gene in this meta-analysis; one SNP in exon 4 of KIAA0319: rs4504469; two SNPs in intron 1of KIAA0319: rs6935076 and rs761100; two SNPs located in the promoter region of KIAA0319 gene: rs9461045 and rs3212236; one SNP in intron 2 of the TTRAP gene: rs2143340.
Exons are represented by numbered grey rectangles. Black parts represent gene’s upstream, introns or downstream.
Results
Characteristics of included studies
The literature search and study selection procedures are shown in Supplementary Fig. S1. After comprehensive searching, 40 potentially relevant reports were retrieved; of which, 11 reports met the inclusion criteria. However, two studies reported by Cope et al. in ref. 23 and Venkatesh et al. in ref. 38 were excluded since the cases largely overlapped with the samples in an analysis by Harold et al. in ref. 24 and Venkatesh et al. in ref. 27, respectively. Although reports by Elbert et al.39 and Couto et al.19 applied the same samples, they focused on different polymorphisms in KIAA0319. In addition, the Eicher et al.21 study’s author kindly replied to our email about asking for original data, and so we also included their data for this meta-analysis21. Thus, ten studies were ultimately eligible for this meta-analysis19,21,24,25,27,28,39,40,41,42. Because one report40 applied two different case groups (dyslexia probands and independent dyslexia) with the same set of controls, we considered it as two case-control studies. Similarly, the Eicher et al.21 study that contained two DD cohorts was considered as two TDT studies. Table 1 shows the characteristics of the included studies. Finally, there are seven case-control studies with 2,711 cases and 2,991 controls and five TDT studies with 943 families from ten reports included in our meta-analysis.
Table 1. Characteristics of included studies.
| Study | Publication year | Study population | Design type | Case/control (family) | KIAA0319 marker |
|---|---|---|---|---|---|
| Harold D | 2006 | Cardiff of UK | Case/control | 350/273 | rs761100, rs4504469, rs3212236, rs6935076, rs2143340 |
| Brkanac Z | 2007 | Washington | TDT | 191 | rs4504469, rs6935076 |
| Couto JM | 2010 | Toronto | TDT | 156 | rs4504469, rs6935076, rs761100, rs2143340 |
| Newbury DF# | 2011 | UK | Case/control | 188/363 | rs9461045, rs761100, rs4504469, rs3212236, rs2143340 |
| Newbury DF* | 2011 | UK | Case/control | 331/363 | rs9461045, rs761100, rs4504469, rs3212236, rs2143340 |
| Elbert A | 2011 | Toronto | TDT | 156 | rs9461045, rs3212236 |
| Kirsten H | 2011 | German | case–control | 272/548 | rs9461045 |
| Venkatesh SK | 2013 | Indian | Case/control | 210/256 | rs9461045, rs4504469, rs3212236 |
| Becker J | 2014 | Eight European countries | Case/control | 958/1150 | rs9461045, rs761100, rs4504469, rs3212236, rs6935076, rs2143340 |
| Eicher JD$ | 2014 | Colorado | TDT | 292 | rs3212236 |
| Eicher JD& | 2014 | Italy | TDT | 304 | rs3212236 |
| Shao SS | 2015 | Chinese | Case/control | 402/401 | rs9461045, rs4504469 |
*Case group is composed of independent dyslexia cases;
#Case group is composed of dyslexia probands;
$ and & represent two independent cohorts from Colorado and Italy, respectively.
Combining the results of case-control and TDT studies
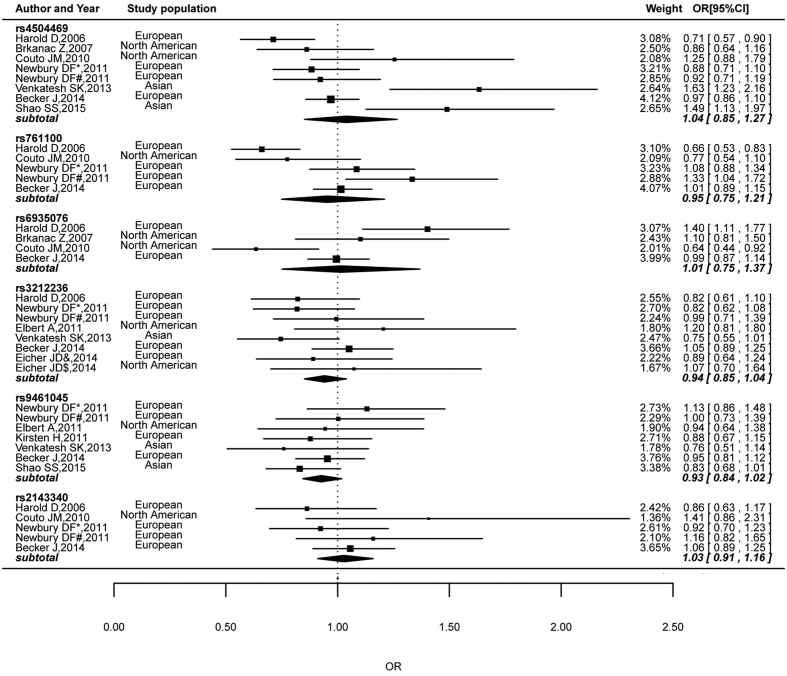
Figure 2 shows the combined results of case-control and TDT studies for six markers in or near the KIAA0319 gene in association with DD risk. Significant heterogeneity was found for rs4504469, rs761100 and rs6935076 (shown in Table 2); thus, the random-effects models were employed for these three markers and the fixed-effects models were applied for the other markers. In the overall meta-analysis, no statistical evidence of associations was found for any included markers and DD risk, with ORs being equal to 1.04 (95% CI = 0.85–1.27, P = 0.706) for rs4504469, 0.95 (95% CI = 0.75–1.21, P = 0.689) for rs761100, 1.01 (95% CI = 0.75–1.37, P = 0.930) for rs6935076, 0.94 (95% CI = 0.85–1.04, P = 0.233) for rs3212236, 0.93 (95% CI = 0.84–1.02, P = 0.114) for rs9461045, and 1.03 (95% CI = 0.91–1.16, P = 0.653) for rs2143340.
Figure 2. Summary of estimated risk of developmental dyslexia associated with six markers in or near the KIAA0319 gene based on meta-analysis.
Table 2. Meta-analysis for associations between KIAA0319 polymorphisms and DD.
| KIAA0319 markers | No.* | Study population | MAF in controls# | Sample sizecases/controls (families) | OR (95% CI) | P value | Heterogeneity |
Model for meta-analysis | Egger’s test |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Q statistic (P value) | I2 (%) | z | P value | ||||||||
| rs4504469 | 8 | Total | 0.13~0.43 | 2439/2443 and 347 | 1.04 (0.85–1.27) | 0.706 | 32.375 (<0.001) | 82.27 | Random | 1.034 | 0.301 |
| 4 | European | 0.40~0.43 | 1827/1786 | 0.90 (0.83–0.99) | 0.028 | 5.427 (0.143) | 45.11 | Fix | −1.552 | 0.121 | |
| 2 | Asian | 0.13, 0.26 | 612/657 | 1.56 (1.28–1.90) | <0.001 | 0.210 (0.647) | 0.00 | Fix | – | – | |
| 2 | North American | – | 347 | 1.03 (0.71–1.48) | 0.889 | 2.535 (0.111) | 60.55 | Fix | – | – | |
| rs761100 | 5 | Total | 0.39~0.46 | 1827/1786 and 156 | 0.95 (0.75–1.21) | 0.689 | 20.002 (<0.001) | 83.28 | Random | −0.472 | 0.637 |
| 4 | European | 0.39~0.46 | 1827/1786 | 0.99 (0.75–1.31) | 0.951 | 18.207 (<0.001) | 87.18 | Random | 0.089 | 0.929 | |
| 1 | North American | – | 156 | 0.78 (0.55–1.10) | 0.155 | – | – | – | – | – | |
| rs6935076 | 4 | Total | 0.35, 0.36 | 1308/1423 and 347 | 1.01 (0.75–1.37) | 0.930 | 13.957 (0.003) | 83.65 | Random | −0.794 | 0.427 |
| 2 | European | 0.35, 0.36 | 1308/1423 | 1.17 (0.83–1.63) | 0.371 | 6.273 (0.012) | 84.06 | Random | – | – | |
| 2 | North American | – | 347 | 0.85 (0.49–1.45) | 0.540 | 5.135 (0.023) | 80.53 | Random | – | – | |
| rs3212236 | 8 | Total | 0.17~0.35 | 2037/2042 and 752 | 0.94 (0.85–1.04) | 0.233 | 7.812 (0.349) | 17.94 | Fix | −0.433 | 0.665 |
| 5 | European | 0.17~0.19 | 1827/1786 and 304 | 0.95 (0.84–1.06) | 0.327 | 3.675 (0.452) | 15.81 | Fix | −1.324 | 0.185 | |
| 1 | Asian | 0.35 | 210/256 | 0.75 (0.55–1.01) | 0.056 | – | – | – | – | – | |
| 2 | North American | – | 448 | 1.14 (0.85–1.53) | 0.375 | 0.150 (0.698) | 0.00 | Fix | – | – | |
| rs9461045 | 7 | Total | 0.18~0.42 | 2361/2718 and 156 | 0.93 (0.84–1.02) | 0.114 | 4.727 (0.579) | 0.00 | Fix | −0.094 | 0.925 |
| 4 | European | 0.18~0.19 | 1749/2061 | 0.98 (0.87–1.10) | 0.678 | 1.841 (0.606) | 0.00 | Fix | 0.368 | 0.713 | |
| 2 | Asian | 0.39, 0.42 | 612/657 | 0.82 (0.68–0.98) | 0.026 | 0.154 (0.695) | 0.00 | Fix | – | – | |
| 1 | North American | – | 156 | 0.94 (0.64–1.39) | 0.770 | – | – | – | – | – | |
| rs2143340 | 5 | Total | 0.15~0.16 | 1827/1786 and 156 | 1.03 (0.91–1.16) | 0.653 | 3.898 (0.420) | 0.01 | Fix | 0.489 | 0.625 |
| 4 | European | 0.15~0.16 | 1827/1786 | 1.01 (0.89–1.14) | 0.903 | 2.243 (0.523) | 0.00 | Fix | −0.479 | 0.632 | |
| 1 | North American | – | 156 | 1.41 (0.86–2.31) | 0.175 | – | – | – | – | – | |
*The number of studies.
#The range of minor allele frequency (MAF) in the control group of the included studies.
Stratified analysis
The data were stratified into three subgroups (European, Asian and North American) according to the study population. The results are shown in Table 2. In the stratified analysis, the heterogeneity for rs761100 and rs6935076 remained significant within each subgroup; the heterogeneity for rs4504469 disappeared, and the study population can explain 86.67% of the variance in findings for rs4504469. The stratified analysis showed that rs4504469 T allele had a protective effect with respect to DD in Europeans (OR = 0.90, 95% CI = 0.83–0.99, P = 0.028), but in Asians the T allele conferred an increased risk of DD (OR = 1.56, 95% CI = 1.28–1.90, P < 0.001). The stratified analysis also revealed that the rs9461045 minor allele (T allele) had a protective effect in the Asian subgroup (OR = 0.82, 95% CI = 0.68–0.98, P = 0.026). The other four markers, rs761100, rs6935076, rs3212236 and rs2143340, were all negatively associated with DD risk in all subgroups.
Given that the analysis stratified by the study population explains the opposite direction of effects for rs4504469 but not the rest of the analyzed SNPs, and that this rs4504469 is located further in the 3′ of the gene (as opposed to the others that were clustered around exon1 as shown in Fig. 1), we decided to assess the differences in the LD pattern of different populations for this SNP using snap: https://www.broadinstitute.org/mpg/snap. The results are shown in Fig. 3.
Figure 3. Regional linkage disequilibrium plot for SNP rs4504469 in CEU and JPT + CHB population panels.
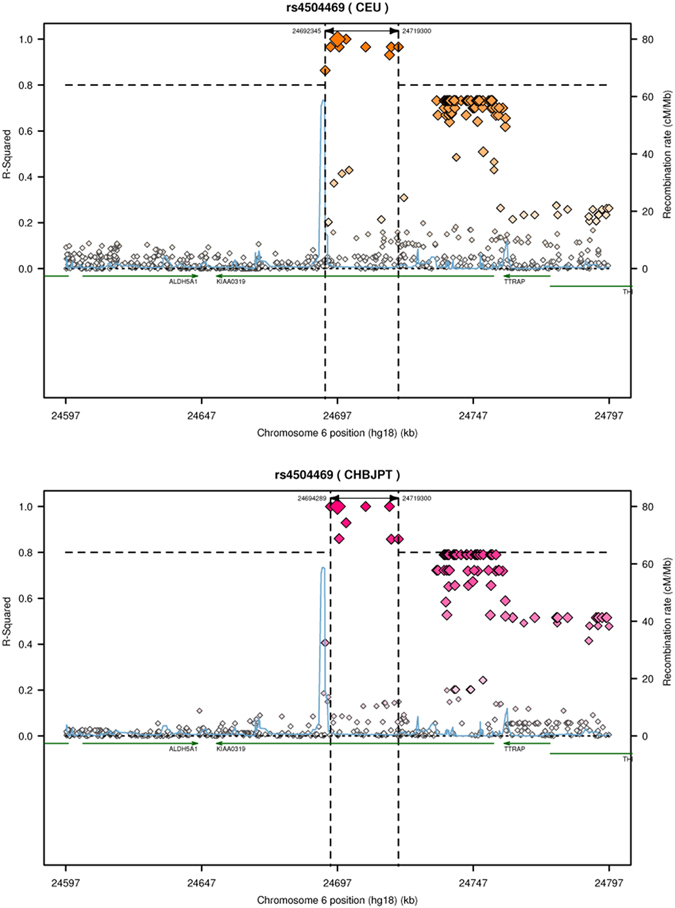
CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; JPT + CHB: Combined panel of Japanese in Tokyo, Japan and Han Chinese in Beijing, China.
Sensitivity analyses for combined studies of KIAA0319 polymorphisms
Given the significant between-study heterogeneity for KIAA0319 rs4504469, rs761100, and rs6935076 polymorphisms, we conducted a sensitivity meta-analysis to assess the effect of each individual study on the combined ORs for these three markers. Random-effect models were employed since heterogeneity was indicated. The results indicated that whichever studies we omitted the heterogeneity in still existed and the associations were still negative for these three markers. Details are shown in Table 3.
Table 3. Sensitivity analysis of pooled ORs for KIAA0319 polymorphisms and DD.
| Study omitted | OR(95% CI) | P | P for heterogeneity | I2 |
|---|---|---|---|---|
| For rs4504469 | ||||
| Harold D, 2006 | 1.10 (0.91–1.33) | 0.347 | 0.001 | 77.85 |
| Brkanac Z, 2007 | 1.07 (0.86–1.33) | 0.570 | 0.000 | 84.56 |
| Couto JM, 2010 | 1.02 (0.82–1.26) | 0.882 | 0.000 | 84.73 |
| Newbury DF*, 2011 | 1.07 (0.85–1.34) | 0.576 | 0.000 | 83.63 |
| Newbury DF#, 2011 | 1.06 (0.84–1.33) | 0.624 | 0.000 | 84.75 |
| Venkatesh SK, 2013 | 0.97 (0.82–1.16) | 0.754 | 0.004 | 74.21 |
| Becker J, 2014 | 1.05 (0.83–1.33) | 0.664 | 0.000 | 81.25 |
| Shao SS, 2015 | 0.99 (0.81–1.20) | 0.897 | 0.001 | 79.73 |
| For rs761100 | ||||
| Harold D, 2006 | 1.05 (0.89–1.24) | 0.531 | 0.082 | 55.63 |
| Couto JM, 2010 | 0.99 (0.75–1.31) | 0.951 | 0.000 | 87.18 |
| Newbury DF*, 2011 | 0.92 (0.68–1.24) | 0.583 | 0.000 | 86.18 |
| Newbury DF#, 2011 | 0.88 (0.70–1.11) | 0.286 | 0.004 | 78.69 |
| Becker J, 2014 | 0.93 (0.68–1.28) | 0.668 | 0.000 | 83.87 |
| For rs6935076 | ||||
| Harold D, 2006 | 0.91 (0.68–1.22) | 0.524 | 0.051 | 72.22 |
| Brkanac Z, 2007 | 0.98 (0.64–1.50) | 0.922 | 0.001 | 90.26 |
| Couto JM, 2010 | 1.14 (0.92–1.42) | 0.232 | 0.043 | 65.97 |
| Becker J, 2014 | 1.01 (0.64–1.59) | 0.962 | 0.002 | 85.56 |
*Case group is composed of independent dyslexia cases.
#Case group is composed of dyslexia probands.
Publication Bias
Egger’s test was performed to assess the publication bias of the literature. No publication bias was detected for any of the included markers. All the P values for Egger’s test were above 0.05 (shown in Table 2).
Power Analysis
Assuming the odds ratio ranges from 1.15 to 1.35 and the standard deviation is 0.05, the power curve is shown in Supplementary Fig. S2. That means, it can detect the odds ratio greater than or equal to 1.15 at power greater than 0.8. Accordingly, this power curve corresponds to the case with odds ratio 1/1.35 −1/1.15, which means odds ranges 0.74–0.87. That is, it can detect the odds ratio smaller than or equal to 0.87 at power greater than 0.8.
Discussion
This meta-analysis of associations between six markers in or near the KIAA0319 gene (rs4504469, rs761100, rs6935076, rs3212236, rs9461045 and rs2143340) and DD risk was based on seven case-control studies comprising a total of 2,711 cases and 2,991 controls and five TDT studies involving 943 families. The results of pooled analysis demonstrated that none of the six markers was associated with DD. A stratified analysis with the study population as the stratification factor revealed that KIAA0319 rs4504469 had opposite associations with DD risk in European and Asian subgroups. The stratified analysis also revealed that the KIAA0319 rs9461045 minor allele had a protective effect in the Asian subgroup, but was a risk allele for UK samples37. This meta-analysis has allowed us to establish the effects of a specific KIAA0319 polymorphism on DD risk with greater precision, revealing that they vary with the study population.
Previously published evaluations of the influence of haplotypes in the KIAA0319 gene on the risk of DD also showed opposite association trends in different samples. Specifically, although under-transmitted haplotypes of rs4504469–rs6935076 were associated with DD in the samples from Cardiff23 and Toronto19, opposite alleles of these markers were involved in the two samples (2-1 in Cardiff and 1-2 in Toronto- where 1 is major allele and 2 is minor allele for each SNP). In addition, two previous studies that evaluated the influence of the KIAA0319 gene on reading skills in the general population drew opposite conclusions about the relationship of the 1-1-2 haplotype (rs4504469–rs2038137–rs2143340) to reading performance (positive association in Australia43; negative association in England44). The two sets of haplotypes overlap in both location and their first component marker, rs4504469 (see Fig. 1).
Lin et al. called these opposite findings a ‘flip-flop phenomenon’ and used theoretical modeling to demonstrate that flip-flop patterns of association can occur when the investigated variant is correlated, through interactive effects or linkage disequilibrium, with a causal variant45. In other words, examining the association between a single genetic marker and DD risk without taking into account other genetic markers of DD risk and environmental factors correlated with the target susceptibility marker will yield ambiguous results45. DD is a complex disease caused by the interaction of various environmental and genetic factors46,47. The genetic backgrounds of Asian and Western people are different, and they have long lived in different environments. Besides, as Fig. 3 shows, the region in high LD (r2 > 0.8) with rs4504469 is not captured by the rest of the analyzed SNPs, and the LD patterns for rs4504469 are different in European and Asian populations. We therefore speculate that there is a functional element in this region and that ethnic differences in inter-marker LD in this region account for the opposite effects of rs4504469, but not the rest of the analyzed SNPs. Of course, differences in inter-marker LD across samples may be due to sampling variation45 rather than ethnicity. It is notable that individuals are usually diagnosed with DD if their reading performance falls below a predetermined threshold but there is at present no universally accepted criteria for this threshold and so the existence of subgroups of individuals with DD diagnoses based on different criteria might lead to disparities in the outcomes of association studies48.
Neither rs761100 nor rs6935076 were associated with DD risk in the meta-analysis, but these results should be treated with caution because of the repeated presence of heterogeneity suggested by the sensitivity analysis and the stratified analysis. Although we do not know the source of the heterogeneity, it seems arbitrary to simply assume that it is due to false positives. The two SNPs might be correlated, through interactive effects or linkage disequilibrium, with a causal variant, which causing flip-flop associations between these two SNPs and DD risk as discussed above. For example, Ludwig et al. found that SNP rs761100 interacted with DCDC2 haplotype rs793862−rs807701 to affect the quantitative subdimension ‘word reading’49 which is the core phenotype for DD in Harold et al. study24.
The other three SNPs that were included in this meta-analysis because of their potential effect on KIAA0319 expression, rs9461045, rs3212236 and rs2143340, were not associated with DD as a discrete trait in each included association studies or in the pooled analysis (see Fig. 2). However, many of the reported associations involving them relate to quantitative reading-related traits24,37,43. Both rs9461045 and rs3212236 are located at the transcription factor-binding site in the promoter region projected by SNP function prediction databases (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm). Rs2143340 is located in the TTRAP gene, which encodes a tumor necrosis factor receptor-associated protein which inhibits nuclear factor-kappa B (NF-κB) transcription50. NF-κB transcription is important for long-term potentiation and synaptic plasticity which are associated with learning and memory23. Based on this we suggest that these three SNPs might be implicated in specific reading skills rather than general reading disability. If possible, a systematic review of research on the effect of KIAA0319 markers on quantitative reading-related traits should be priority for future research.
In summary, the meta-analysis clarified the associations between KIAA0319 polymorphisms and DD risk and suggested that the associations between single SNPs and DD risk vary according to the study population. There may be multiple genetic variants (rather than a single SNP) that contribute to DD risk. Further primary research and updated meta-analysis of findings on KIAA0319 is needed to explore the combined effect of multiple genetic polymorphisms and environmental factors on DD risk. In complex diseases the effect of single causal variant is very weak, and the sample sizes in our analysis were relatively small, especially in the stratified analysis. More studies with larger samples in a particular ethnic group are needed to confirm our findings and identify true causal variants via fine-mapping.
Methods
Search strategy and identification of relevant studies
We searched PubMed, EMBASE, and ISI Web of Science databases for published articles up to May 2015 using the keywords “KIAA0319”, “dyslexia or reading disability”, “polymorphism or variant” without language restrictions. References of the retrieved articles were also scanned. Reviews, comments, and letters were also checked for additional studies.
The following inclusion criteria had to be fulfilled: (1) either case-control or TDT study design; (2) KIAA0319 polymorphisms and DD risk; (3) allele frequencies on KIAA0319 polymorphisms in case and control groups for case–control studies, and numbers of transmitted alleles from heterozygous parents to affected offspring for family-based studies. For the study that only meet the first two conditions, we emailed its corresponding author to asking for the original data mentioned in the third condition. Animal studies, reviews, simple commentaries and meetings were excluded. Study overlap was eliminated by selecting the report with a complete design or larger sample size.
Data extraction
All data were extracted in dependently by two reviewers (S. Shao & X. Zhang). The following information was extracted from the eligible studies: first author’s name, year of publication, study population, study design, and KIAA0319 polymorphisms. The counts of alleles in case and control groups in case-control studies and the number of transmitted alleles from heterozygous parents to affected offspring in family-based studies were extracted or calculated in the included studies.
Statistical analysis
All statistical analyses were conducted using the metafor package (version 1.9-5) in R (version 3.2.3; http://www.r-project.org/). Briefly, data from each case-control and TDT study were extracted and sorted as per the metafor package instructions. Odds ratios (ORs), 95% confidence intervals (95%CIs) and standard errors (SEs) were calculated for individual study based on allele data. The between-study heterogeneity was assessed by a χ2-based Cochran’s Q-statistic test (the heterogeneity was considered significant at P < 0.10). A fixed-effects model was used to apply data from studies when heterogeneity was negligible; otherwise, a random-effects model was applied. For the synthesis of case-control and TDT studies, Kazeem and Farrall51 outlined a methodological improvement for achieving integration by a fixed-effects approach, and then Nicodemus subsequently extended this method to the random-effects model52. Stratified analyses were conducted according to the study population. Publication bias was assessed by Egger’s test53. In addition, a sensitivity analysis was performed to evaluate the influence of each study on the overall estimate. The power is calculated based on power formula of the mixed model for individual regression coefficients54. All P values were two-tailed with a significance level at 0.05, except for Cochran’s Q-statistic test.
Additional Information
How to cite this article: Shao, S. et al. Opposite Associations between Individual KIAA0319 Polymorphisms and Developmental Dyslexia Risk across Populations: A Stratified Meta-Analysis by the Study Population. Sci. Rep. 6, 30454; doi: 10.1038/srep30454 (2016).
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China NSFC-81273092; the Fundamental Research Funds for the Central Universities (HUST: 2014TS053, 2015TS096); Project from Health and Family Planning Commission of Hubei Province (WJ2015MB019).
Footnotes
Author Contributions S.S., Y.N., J.Z. and R.S. designed the research; S.S., Y.N. and R.S. wrote the paper; S.S. and X.Z. reviewed and extracted data from eligible studies; X.Z., R.K., J.Z. and R.S. performed the statistical analysis; X.Z., J.W., L.L. and X.L. prepared the tables and figures. All authors have reviewed and approved the manuscript as submitted.
References
- Peterson R. L. & Pennington B. F. Developmental dyslexia. J. Lancet. 379, 1997–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. S.-H., Chan D. W.-O., Lee S.-H., Tsang S.-M. & Luan V. H. Cognitive profiling and preliminary subtyping in Chinese developmental dyslexia. J. Cognition. 91, 43–75 (2004). [DOI] [PubMed] [Google Scholar]
- Chung K. K. et al. The role of visual and auditory temporal processing for Chinese children with developmental dyslexia. J. Ann Dyslexia. 58, 15–35 (2008). [DOI] [PubMed] [Google Scholar]
- Chung K. K., Ho C. S. H., Chan D. W., Tsang S. M. & Lee S. H. Cognitive profiles of Chinese adolescents with dyslexia. J. Dyslexia. 16, 2–23 (2010). [DOI] [PubMed] [Google Scholar]
- Francis D. J., Shaywitz S. E., Stuebing K. K., Shaywitz B. A. & Fletcher J. M. Developmental lag versus deficit models of reading disability: A longitudinal, individual growth curves analysis. J. Educ Psychol. 88, 3–17 (1996). [Google Scholar]
- Shaywitz S. E. & Shaywitz B. A. Dyslexia (specific reading disability). J. Biol Psychiatry. 57, 1301–1309 (2005). [DOI] [PubMed] [Google Scholar]
- Ritchie S. J. & Bates T. C. Enduring links from childhood mathematics and reading achievement to adult socioeconomic status. J. Psychol Sci. 24, 1301–1308 (2013). [DOI] [PubMed] [Google Scholar]
- Richardson J. T. & Wydell T. N. The representation and attainment of students with dyslexia in UK higher education. J. Read Writ. 16, 475–503 (2003). [Google Scholar]
- He Z. et al. Does long time spending on the electronic devices affect the reading abilities? A cross-sectional study among Chinese school-aged children. J. Res Dev Disabil. 35, 3645–3654 (2014). [DOI] [PubMed] [Google Scholar]
- Davis O. S. et al. The correlation between reading and mathematics ability at age twelve has a substantial genetic component. J. Nat Commun. 5, 4204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko E. L. Genetic bases of developmental dyslexia: A capsule review of heritability estimates. J. Enfance. 56, 273–288 (2004). [Google Scholar]
- Williams J. & O’Donovan M. C. The genetics of developmental dyslexia. J. Eur J Hum Genet. 14, 681–689 (2006). [DOI] [PubMed] [Google Scholar]
- Kaplan D. E. et al. Evidence for linkage and association with reading disability on 6p21.3-22. J. Am J Hum Genet. 70, 1287–1298 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayan J. et al. Quantitative-trait locus for specific language and reading deficits on chromosome 6p. J. Am J Hum Genet. 64, 157–164 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko E. L., Wood F. B., Meyer M. S. & Pauls D. L. Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. J. Am J Hum Genet. 66, 715–723 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francks C. et al. A 77-kilobase region of chromosome 6p22. 2 is associated with dyslexia in families from the United Kingdom and from the United States. J. Am J Hum Genet. 75, 1046–1058 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini S. et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. J. Hum Mol Genet. 15, 1659–1666 (2006). [DOI] [PubMed] [Google Scholar]
- Ludwig K. U. et al. Investigation of interaction between DCDC2 and KIAA0319 in a large German dyslexia sample. J. J Neural Transm. 115, 1587–1589 (2008). [DOI] [PubMed] [Google Scholar]
- Couto J. M. et al. Association of reading disabilities with regions marked by acetylated H3 histones in KIAA0319. J. Am J Med Genet B Neuropsychiatr Genet. 153B, 447–462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope N. et al. Variants in the DYX2 locus are associated with altered brain activation in reading-related brain regions in subjects with reading disability. J. NeuroImage. 63, 148–156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher J. D. et al. Characterization of the DYX2 locus on chromosome 6p22 with reading disability, language impairment, and IQ. J. Hum Genet. 133, 869–881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scerri T. S. et al. DCDC2, KIAA0319 and CMIP are associated with reading-related traits. J. Biol Psychiatry. 70, 237–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope N. et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. J. Am J Hum Genet. 76, 581–591 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D. et al. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. J. Mol Psychiatr. 11, 1085–1091 (2006). [DOI] [PubMed] [Google Scholar]
- Brkanac Z. et al. Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. J. Am J Med Genet B Neuropsychiatr Genet. 144, 556–560 (2007). [DOI] [PubMed] [Google Scholar]
- Zou L. et al. Genetic variant in KIAA0319, but not in DYX1C1, is associated with risk of dyslexia: an integrated meta-analysis. J. Am J Med Genet B Neuropsychiatr Genet. 159B, 970–976 (2012). [DOI] [PubMed] [Google Scholar]
- Venkatesh S. K., Siddaiah A., Padakannaya P. & Ramachandra N. B. Analysis of genetic variants of dyslexia candidate genes KIAA0319 and DCDC2 in Indian population. J. J Hum Genet. 58, 531–538 (2013). [DOI] [PubMed] [Google Scholar]
- Shao S. et al. The Roles of Genes in the Neuronal Migration and Neurite Outgrowth Network in Developmental Dyslexia: Single- and Multiple-Risk Genetic Variants. J. Mol Neurobiol. doi: 10.1007/s12035-015-9334-8 (2015). [DOI] [PubMed] [Google Scholar]
- Galaburda A. M., LoTurco J., Ramus F., Fitch R. H. & Rosen G. D. From genes to behavior in developmental dyslexia. J. Nat Neurosci. 9, 1213–1217 (2006). [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O. et al. Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. J. Hum Mol Genet. 9, 1641–1649 (2000). [DOI] [PubMed] [Google Scholar]
- Levecque C., Velayos-Baeza A., Holloway Z. G. & Monaco A. P. The dyslexia-associated protein KIAA0319 interacts with adaptor protein 2 and follows the classical clathrin-mediated endocytosis pathway. J. Am J Physiol Cell Physiol. 297, C160–168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos-Baeza A., Toma C., Paracchini S. & Monaco A. P. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. J. Hum Mol Genet. 17, 859–871 (2008). [DOI] [PubMed] [Google Scholar]
- Szalkowski C. E. et al. Neocortical disruption and behavioral impairments in rats following in utero RNAi of candidate dyslexia risk gene Kiaa0319. J. Int J Dev Neurosci. 30, 293–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanni T. M. et al. Speech sound processing deficits and training-induced neural plasticity in rats with dyslexia gene knockdown. J. PloS one. 9, e98439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda A. M. Dyslexia—A molecular disorder of neuronal migration. J. Ann Dyslexia. 55, 151–165 (2005). [DOI] [PubMed] [Google Scholar]
- Mascheretti S. et al. KIAA0319 and ROBO1: evidence on association with reading and pleiotropic effects on language and mathematics abilities in developmental dyslexia. J. J Hum Genet. 59, 189–197 (2014). [DOI] [PubMed] [Google Scholar]
- Dennis M. Y. et al. A common variant associated with dyslexia reduces expression of the KIAA0319 gene. J. PLoS Genet. 5, e1000436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S. K., Siddaiah A., Padakannaya P. & Ramachandra N. B. An examination of candidate gene SNPs for dyslexia in an Indian sample. J. Behav Genet. 41, 105–109 (2011). [DOI] [PubMed] [Google Scholar]
- Elbert A. et al. Genetic variation in the KIAA0319 5’ region as a possible contributor to dyslexia. J. Behav Genet. 41, 77–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury D. F. et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. J. Behav Genet. 41, 90–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsten H., Wilcke A., Ligges C., Boltze J. & Ahnert P. Association study of a functional genetic variant in KIAA0319 in German dyslexics. J. Psychiat Genet. 22, 216–217 (2012). [DOI] [PubMed] [Google Scholar]
- Becker J. et al. Genetic analysis of dyslexia candidate genes in the European cross-linguistic NeuroDys cohort. J. Eur J Hum Genet. 22, 675–680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M. et al. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. J. Biol Psychiatry. 62, 811–817 (2007). [DOI] [PubMed] [Google Scholar]
- Paracchini S. et al. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. J. Am J Psychiatry. 165, 1576–1584 (2008). [DOI] [PubMed] [Google Scholar]
- Lin P. I., Vance J. M., Pericak-Vance M. A. & Martin E. R. No gene is an island: the flip-flop phenomenon. J. Am J Hum Genet. 80, 531–538 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascheretti S., Bureau A., Trezzi V., Giorda R. & Marino C. An assessment of gene-by-gene interactions as a tool to unfold missing heritability in dyslexia. J. Hum Genet. 134, 749–760 (2015). [DOI] [PubMed] [Google Scholar]
- Sun Z. et al. Prevalence and associated risk factors of dyslexic children in a middle-sized city of China: a cross-sectional study. J. PloS one. 8, e56688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini S., Scerri T. & Monaco A. P. The genetic lexicon of dyslexia. J. Annu Rev Genomics Hum Genet. 8, 57–79 (2007). [DOI] [PubMed] [Google Scholar]
- Ludwig K. U. et al. Investigation of the DCDC2 intron 2 deletion/compound short tandem repeat polymorphism in a large German dyslexia sample. J. Psychiat Genet. 18, 310–312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pype S. et al. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. J Biol Chem. 275, 18586–18593 (2000). [DOI] [PubMed] [Google Scholar]
- Kazeem G. R. & Farrall M. Integrating case-control and TDT studies. J. Ann Hum Genet. 69, 329–335 (2005). [DOI] [PubMed] [Google Scholar]
- Nicodemus K. K. Catmap: case-control and TDT meta-analysis package. J. BMC Bioinformatics. 9, 130 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M., Smith G. D., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. J. BMJ. 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges L. V. & Pigott T. D. The Power of Statistical Tests in Meta-Analysis. J. Psychol Methods. 6, 203–217 (2001). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.