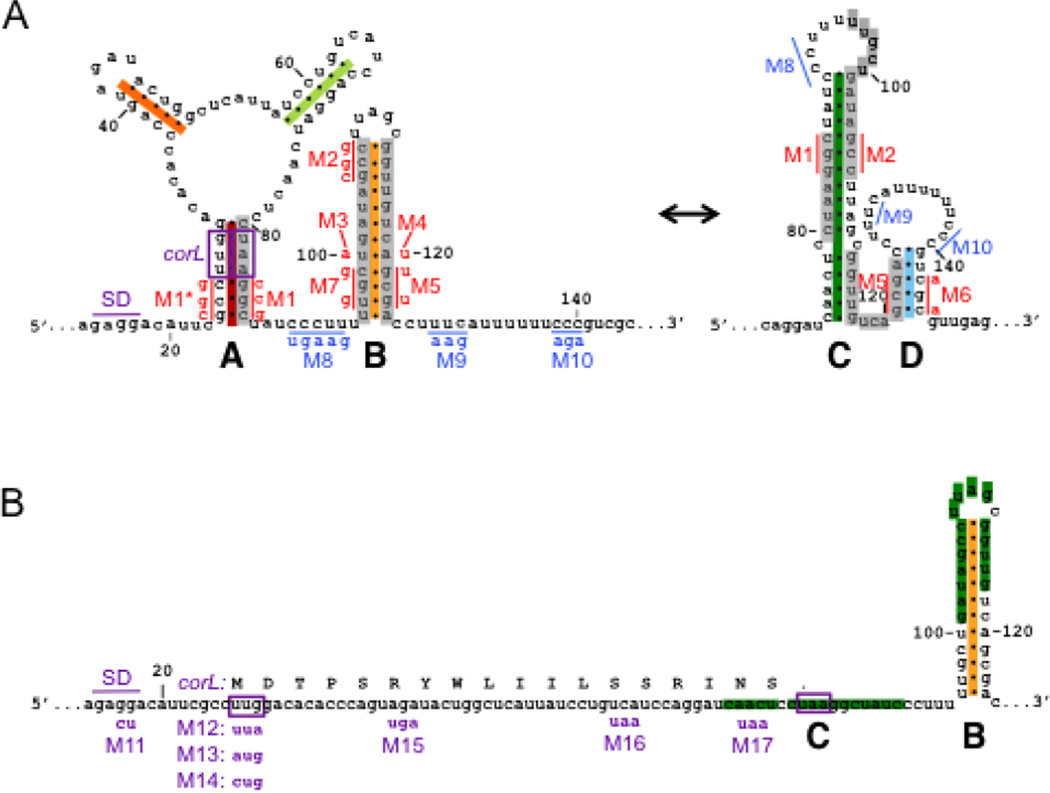
Fig. 3. The corA leader can adopt two mutually exclusive conformations that differentially regulate expression of the associated coding region.
(A) Schematic of predicted secondary structures within the corA leader mRNA. The two possible conformations, termed stem-loops A+B and stem-loops C+D, are mutually exclusive (grey bars). Positions are numbered relative to the corA TSS. Mutations used to disrupt base pairing (red) or Rho loading (blue) are denoted next to the mutated nucleotides. Structure prediction and mutation design were aided by the M-fold Web Server (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form). (B) Schematic denoting the position of the short ORF corL relative to the RNA secondary structures. The corL SD sequence, start codon and stop codon are shown in purple. Translation of corL in vivo is expected to favor stem-loop B by preventing formation of stem-loops A and C. Mutations used to alter corL translation initiation or elongation are denoted below the nucleotide sequence.