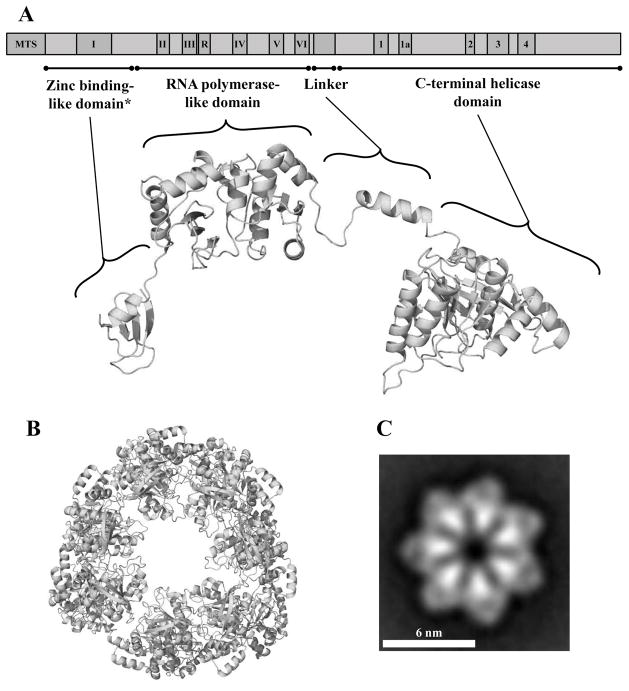
Figure 3.
Schematic representation and structural model of the human mtDNA helicase. A, Upper image, schematic representation of the conserved amino acid sequence motifs in human mtDNA helicase. MTS, mitochondrial target sequence; I–VI and R, conserved sequence motifs I–IV and RNAP basic motif of prokaryotic primases; 1–4 and 1a, conserved sequence motifs 1–4 and 1a of ring-shaped helicases [167]. The size and position of the conserved sequence motifs are represented to scale. Lower image, structural model of a protomer of the human mtDNA helicase highlighting its modular architecture organized in a zinc binding-like domain (ZBD), RNA polymerase-like domain (RPD), Linker region and a C-terminal helicase domain (CTD). * ZBD portion is represented as the polypeptide backbone of the bacteriophage T7 gp4 ZBD. B, Model of the heptameric human mtDNA helicase, CTD view. C, Electron microscopic image of the recombinant human mtDNA helicase at 100 mM NaCl in the presence of Mg2+ and ATPγS, showing its heptameric configuration. Reproduced with permission from “L.S. Kaguni, M.T. Oliveira: Structure, function and evolution of the animal mitochondrial replicative DNA helicase. Critical Reviews in Biochemistry and Molecular Biology (2016) 51, 53–64” and from “T.D. Ziebarth, R. Gonzalez-Soltero, M.M. Makowska-Grzyska, R. Núñez-Ramírez, J.M. Carazo, L.S. Kaguni: Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. Journal of Biological Chemistry (2010) 285, 14639–14647”.