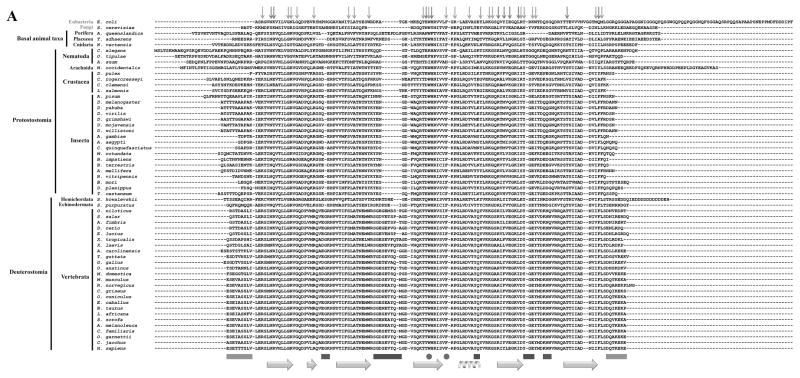
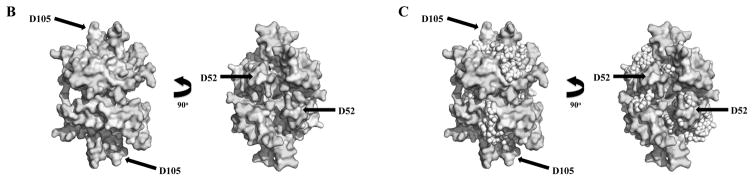
Figure 4.
Evolutionary Trace analysis of animal mtSSBs. A, Multiple amino acid sequence alignment using selected animal mtSSBs retrieved from public databases (NCBI, Ensembl Metazoa and 959 Nematode Genomes). TBLASTN [168] and HMMR3 BLAST [169] searches were performed using the translated mRNA reference sequences for the Homo sapiens (AK313033.1) and Drosophila melanogaster (BT016028.1) mtSSB as queries. The alignment was performed using the MUSCLE algorithm built into the MEGA6 software [170]. The outgroup sequences used here were the Saccharomyces cerevisiae mtSSB (S43128.1) and the Escherichia coli SSB (J01704.1). Red and light green arrows above the alignment indicate the residues that are crucial for ssDNA binding and subunit interactions in the E. coli SSB protein, respectively [171, 172]. Gray, purple and dark green bars below the alignment indicate, respectively, three mtSSB regions grouped by their functional properties: N- and C- termini (which have negative effects on Pol γ stimulation [46]); loop 2,3, alpha-1 and loop 4,5-1 (which have positive effects on Pol γ stimulation [47]); and loop 1,2 and loop 4,5-2 (which have positive effects on mtDNA helicase stimulation [47]). The cyan arrows and helix below the alignment indicate the residues that form the β sheets and the only α-helix in the mtSSB polypeptide, according to the crystal structure (PDB: 3ULL, [59]). The residues highlighted in yellow were identified by the Evolutionary Trace Server [56, 57] as conserved in metazoans. These have been mapped onto the crystal structure of the human mtSSB (B) and onto a model of ssDNA-bound mtSSB (C). The model is as reported by Oliveira and Kaguni [47]. D52 and D105 are the only residues identified by the Evolutionary Trace analysis that are most likely not involved in ssDNA binding (see text for details).