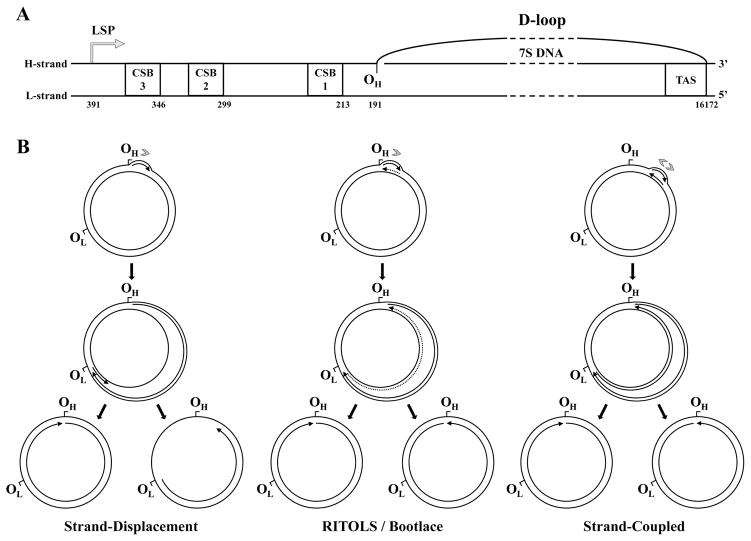
Figure 8.
Current models of vertebrate mtDNA replication. A, Structural organization of the D-loop and the adjacent cis-elements present within the non-coding region of vertebrate mtDNA. CSB1, 2 and 3, conserved sequence blocks 1, 2 and 3; TAS, termination associated sequence; LSP, light strand promoter; OH, origin of heavy strand DNA synthesis. The numbers below each element represent genomic positions in the human mtDNA reference sequence. The schematic is to scale, except for the region represented by the dashed lines. B, Strand-displacement, RITOLS/bootlace and strand-coupled models of vertebrate mtDNA replication (see text for details). The sites OH and OL (origin of light strand DNA synthesis) are represented as reference points on the genome map, although these sites are important primarily for the strand-displacement model. Arrows associated with replicating mtDNA indicate the 5′- to 3′-direction of DNA synthesis; continuous and dotted lines represent DNA and RNA, respectively. Only the long stretches of RNA described in the RITOLS model are represented; the putative short RNA primers of the other models are not shown. Gray arrowheads indicate the number and directionality of replication forks generated at the origin according to each model. Adapted from “E.A. McKinney, M.T. Oliveira: Replicating animal mitochondrial DNA. Genetics and Molecular Biology (2013) 36, 308–315”.