ABSTRACT
Vertebrate genomes contain many virus-related sequences derived from both retroviruses and non-retroviral RNA and DNA viruses. Such non-retroviral RNA sequences are possibly produced by reverse-transcription and integration of viral mRNAs of ancient RNA viruses using retrotransposon machineries. We refer to this process as transcript reversion. During an ancient bornavirus infection, transcript reversion may have left bornavirus-related sequences, known as endogenous bornavirus-like nucleoproteins (EBLNs), in the genome. We have recently demonstrated that all Homo sapiens EBLNs are expressed in at least one tissue. Because species with EBLNs appear relatively protected against infection by a current bornavirus, Borna disease virus, it is speculated that EBLNs play some roles in antiviral immunity, as seen with some endogenous retroviruses. EBLNs can function as dominant negative forms of viral proteins, small RNAs targeting viral sequences, or DNA or RNA elements modulating the gene expression. Growing evidence reveals that various RNA viruses are reverse-transcribed and integrated into the genome of infected cells, suggesting transcript reversion generally occurs during ongoing infection. Considering this, transcript reversion-mediated interference with related viruses may be a novel type of antiviral immunity in vertebrates. Understanding the biological significance of transcript reversion will provide novel insights into host defenses against viral infections.
KEYWORDS: antiviral immunity, endogenous viral element, interference, LINE-1, non-coding RNA, piRNA, RNA virus, transcript reversion
Endogenous bornavirus-like nucleoproteins: Fossil records of ancient bornavirus infection
It is well known that eukaryotic genomes contain many virus-related sequences, mostly derived from retroviruses. In addition to retroviruses, we and others have recently discovered that numerous vertebrate genomes contain endogenous sequences derived from non-retroviral RNA and DNA viruses.1-3 Bornavirus, a nonsegmented, negative-strand RNA virus, is unique because it is the only non-retroviral RNA virus with endogenous elements found embedded in the human genome.1 The majority of endogenous fragments of bornavirus in human genomes appear to have originated from the reverse-transcription and integration of nucleoprotein (N) mRNA of ancient bornavirus via long interspersed nuclear element-1 (LINE-1, L1) activity (Fig. 1).1 We call these sequences, endogenous bornavirus-like nucleoproteins (EBLNs).1 EBLNs are thus evidence of a novel mechanism of retrotransposon-mediated RNA-to-DNA information flow from virus to host. We refer to this retrotransposon-mediated information flow as transcript reversion. Because species with EBLNs appear relatively protected against a current bornavirus, Borna disease virus (BDV), it is speculated that EBLNs play some roles in antiviral immunity in a similar manner to those of several endogenous retroviruses (ERVs).2 In this commentary, we will introduce some proposed functions of EBLNs in antiviral immunity.
Figure 1.
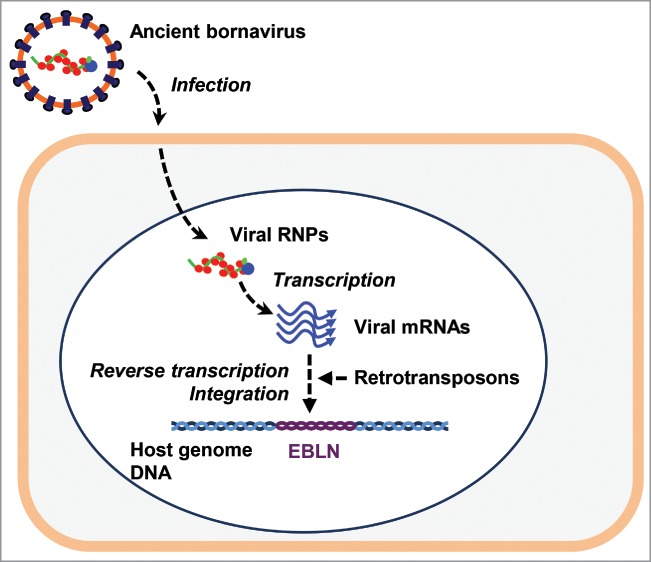
Transcript reversion during bornavirus infection. After infecting host cells, bornavirus transcribes its mRNAs using bornavirus RNP as a template. L1 reverse-transcribes viral mRNAs into viral cDNAs and integrates them into the genome of infected cells. The cDNAs of ancient bornavirus sequences are endogenized during evolution, forming EBLNs in the genome of a descendant of infected species.
Ictidomys tridecemlineatus EBLN: A dominant negative form of a viral protein
Some EBLNs have conserved their long open reading frames (ORFs) during evolution. For example, 2 EBLNs in the human genome, Homo sapiens EBLN (hsEBLN) -1 and -2, contain long ORFs with the potential to code for proteins of 366 and 225 amino acids, respectively.1 These proteins have an overall 41% amino acid sequence identity to BDV N protein. Among EBLNs in various genomes, one in the thirteen-lined ground squirrel genome, Ictidomys tridecemlineatus EBLN (itEBLN), has an ORF with relatively high amino acid sequence identity (77%) to BDV N protein.4 It has been demonstrated that ERV-derived proteins, such as Friend virus susceptibility 1 (Fv1), protect host cells from infection by exogenous related viruses.5,6 Most known examples of this phenomenon are mediated by the production of a dominant negative form of viral proteins.7 We therefore reasoned that itEBLN also inhibits BDV infection as a dominant negative form of BDV N protein. Recently, we reported that itEBLN co-localizes with a viral factory that consists of BDV ribonucleoprotein complex (RNP).4 Furthermore, itEBLN associates with BDV RNP and inhibits BDV polymerase activity (Fig. 2A).4 Several ERV-derived elements are known to co-assemble with the nucleocapsids of exogenous viruses, inhibiting their infections. In the case of Fv1, an element derived from the gag gene of an ancient retrovirus, this directly interacts with the capsid protein of the murine leukemia virus (MLV) nucleocapsid, inducing anti-MLV immunity.8 An amino acid comparison between itEBLN and N reveals that the identity between itEBLN and N may be sufficient to induce heteromultimerization. On the other hand, a number of substitutions are found in itEBLN sequences. Such observations support the hypothesis that itEBLN co-assembles into viral RNPs and acts in a dominant negative manner.4 However, although itEBLN functions as a dominant negative form of the N protein, this might be a rare case because the ORF of EBLNs has to be conserved, and complex optimal amino acid substitutions are required for gaining such a function.
Figure 2.
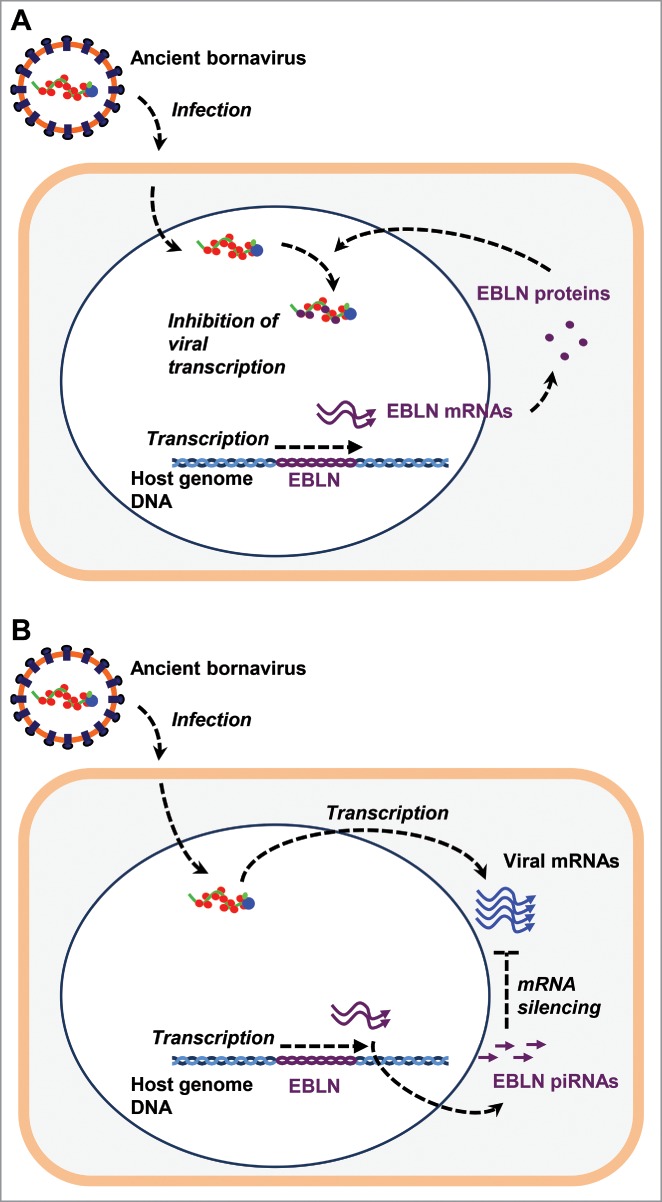
EDI in ancient bornavirus infection. (A) Dominant negative forms of viral proteins. EBLN proteins encoded in EBLN loci can co-assemble into viral RNPs of exogenous related viruses, inhibiting viral transcription. (B) TRAPS. EBLNs within piRNA clusters produce piRNAs. These EBLN-derived piRNAs post-transcriptionally silence viral mRNA of exogenous related viruses.
Mus musculus EBLNs: PIWI-interacting RNAs potentially targeting viral sequences
While itEBLN can inhibit BDV replication,4 most EBLNs in other species have disrupted their ORFs. Furthermore, there is no evidence of selection to maintain the ORF of EBLNs in primates.9 These observations suggest that most EBLNs either had lost their function or had a function not related to protein-coding, such as non-coding RNAs (ncRNAs). We have recently demonstrated that all 7 hsEBLNs are expressed as RNA in at least one tissue.10 Furthermore, we have observed that 4 of the 7 hsEBLNs are currently transcribed fairly ubiquitously.10 This finding is unexpected based on the transcription of pseudogenes processed from host mRNA, of which only 4 to 6 percent are transcribed.11 The unexpectedly large extent of transcription from EBLN loci raises the prospect that EBLNs may encode antiviral RNA. In this regard, we have noticed that some EBLNs in the rodent and primate genomes, such as Mus musculus EBLN (mmEBLN) -3 to -5, are present within PIWI-interacting RNA (piRNA)-generating loci, called “piRNA clusters,” of the genome far more often than expected by chance alone.12 In complex with a PIWI clade argonoute protein, piRNA targets transposons with complimentary sequences for transcriptional and post-transcriptional silencing.13 It has been previously noted that similarities exist between the piRNA pathway and the CRISPR/Cas system, an RNA-mediated, trangenerational antiviral immunity in prokaryotes.14,15 One such resemblance is that piRNAs are transcribed from discrete piRNA clusters similar to CRISPR arrays. Because of the conceptual similarities between these 2 systems, piRNA derived from EBLNs may be an attractive candidate for an EBLN-encoding antiviral RNA. We and others have indeed detected rodent and primate EBLN-derived piRNAs.12,16,17 Notably, these EBLN-derived piRNAs mapped to the antisense strand relative to bornavirus N mRNA, suggesting the potential to post-transcriptionally silence bornavirus mRNA (Fig. 2B).12
While EBLN loci are not syntenic between rodents and primates, some piRNA clusters containing EBLNs are.1,18 We have therefore deduced that piRNA-generating EBLNs have been integrated into existing piRNA clusters.12 The apparent convergent evolution of piRNA-generating EBLNs in both rodents and primates can be explained by the preferential integration of viral sequences within piRNA clusters. Alternatively, viral sequences may have integrated randomly, while those within piRNA clusters may have been selected during species evolution. Because sequences mobilized by L1 machineries are not preferentially targeted to piRNA clusters,19,20 the latter possibility is more likely. In either case, EBLNs in piRNA clusters can produce piRNAs that potentially target bornavirus sequences. Our observations thus raise the hypothesis, referred as viral transcript reversion with anamnestic piRNA silencing (TRAPS), that the host may capture virus sequences in its specialized genome loci to engender an RNA-mediated, sequence-specific, antiviral immune memory.12 Although the confirmed targets of piRNAs are largely limited to endogenous retrotransposons,21 the transgene silencing observed after the insertion of identical sequences into piRNA clusters supports our hypothesis.22 Further studies are required to investigate whether EBLN-derived piRNAs do target bornavirus sequences, in vivo and/or in vitro, to evaluate this hypothesis.
hsEBLN-1: A potential immunomodulatory RNA
Retrotransposons were first characterized as elements capable of controlling the expression of neighboring genes.23 Indeed, ERVs, L1s and processed pseudogenes can influence the expression of neighboring genes by various mechanisms. Furthermore, retrotransposons or transposed genes, as a regulatory DNA, can regulate the expression of homologous genes in other loci by competing for transcription factor(s).24,25 Likewise, EBLN may have the potential to regulate the expression of other genes (Fig. 3). We have recently demonstrated that hsEBLN-1 may affect the expression of its neighboring COMMD3 gene.10 When expression of the EBLN-1 locus in the primate genomes was induced, expression of the COMMD3 gene became downregulated. The effect of induction of hsEBLN-1 RNA on the COMMD3 locus was abrogated by treatment with siRNA against hsEBLN-1 RNA. Although hsEBLN-1 has an ORF encoding 366 amino acids, comparable with the full-length BDV N protein, the involvement of the hsEBLN-1 protein in the COMMD3 gene expression is unlikely because the protein is located in the cytoplasm.4 These results suggest that hsEBLN-1 RNA may function as a long non-coding RNA (lncRNA) that scaffolds transcriptional repressors for the COMMD3 gene around the locus, downregulating its expression.10 We cannot exclude the possibility that hsEBLN-1 functions as a regulatory DNA element (Fig. 3), but our data using siRNA strongly suggest a role for hsEBLN-1 as a lncRNA. Because the COMMD3 gene encodes a protein that can interact with and inhibit the NFκB pathway,26 EBLN insertion in this locus may downregulate the expression of the COMMD3 gene and thereby potentiate the NFκB pathway of host cells to counteract invading pathogens. Further studies are required for understanding the contribution of EBLNs to immune modulation.
Figure 3.
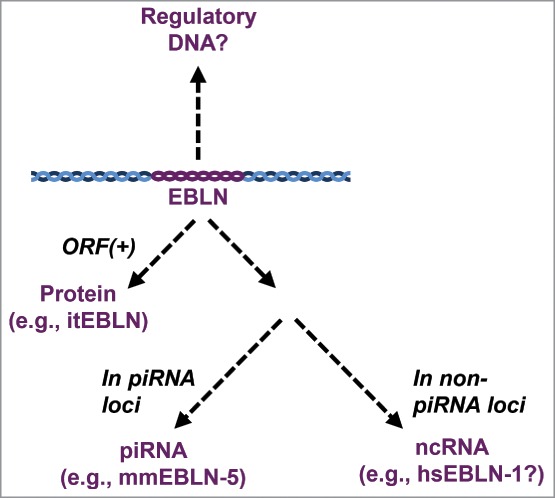
Categorization of EBLN characteristics and functions. The functional mechanisms of EBLNs can be categorized in terms of their characteristics as follows: a regulatory DNA, a functional protein, and a functional ncRNA. All EBLNs can potentially regulate surrounding genes. EBLNs with ORFs may function as functional proteins, such as the case of itEBLN acting as a dominant negative protein. If EBLNs locate within piRNA clusters, these may produce piRNA targeting related exogenous viruses (e.g., mmEBLN-5). Even if EBLNs do not contain ORFs and locate within piRNA clusters, EBLNs can still function as ncRNAs as proposed for hsEBLN-1.
Transcript reversion-mediated interference
EBLNs provides evidence that endogenous viral element-derived immunity (EDI) is found not only in endogenous retroviruses, but also in endogenous non-retroviral virus elements.4,7,10 Beyond this, EBLN-mediated EDI may add to the concept of a novel, antiviral response against ongoing infection. Theoretically, transcript reversion mediated by L1 can occur for all viruses, including RNA and DNA viruses. Retrotransposons, other than L1, could also mediate transcript reversion of virus sequences, such as is the case for lymphocytic choriomeningitis virus (LCMV).27 Indeed, growing evidence reveals that various types of RNA viruses are reverse-transcribed and integrated into the genome of infected cells, suggesting that transcript reversion is a general phenomenon found in host cells during ongoing infection.1-3,28-32 Given that transcript reversion occurs in cells infected with various types of viruses, virus-derived integrants in the infected cells can function similarly to EDI before endogenization of the sequences. Although no such activity has been ascribed to virus-derived sequences in mammals, a similar, virus-derived sequence can control the infection of an RNA virus in Drosophila.33 We therefore propose that integrants derived from infected, non-retroviral RNA viruses may confer antiviral immunity resembling EDI in the infected cells. We distinguish this novel type of antiviral immunity from EDI conferred by endogenized viral elements and refer to it as transcript reversion-mediated interference (TRi). TRi provides 2 unique concepts regarding antiviral immunity. Firstly, of the EDI mechanisms for EBLNs described above, RNA-mediated mechanisms, such as TRAPS and immunomodulatory lncRNAs, could be candidate mechanisms of TRi in infected cells. This is because the only requirement for an RNA-mediated TRi is the integration of viral sequences into discrete loci downstream of an existing promoter, such as piRNA clusters. Reports of such RNA-mediated immunity in metazoans are limited so far. Another interesting point in regard to TRi is that TRi could provide a transgenerational antiviral immunity in prokaryotes. Although further studies are clearly required to evaluate this hypothesis, understanding the biological significance of transcript reversion will provide novel insights into host defenses against various viral infections.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported in part by KAKENHI Grant Numbers 26253027, 26670225 and 15H01259 (KT), and 24115709, 25115508, 25860336 and 15K08496 (TH) from Japan Society for the Promotion of Science (JSPS), a Basic Science and Platform Technology Program for Innovative Biological Medicine (KT) from Japan Agency for Medical Research and Development (AMED), and grants from the Takeda Science Foundation, Senri Life Science Foundation, Suzuken Memorial Foundation, The Shimizu Foundation for Immunology and Neuroscience Grant for 2015 and The NOVARTIS Foundation (Japan) for the Promotion of Science (TH).
References
- [1].Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, et al.. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010; 463:84-7; PMID:20054395; http://dx.doi.org/ 10.1038/nature08695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog 2010; 6:e1001030; PMID:20686665; http://dx.doi.org/ 10.1371/journal.ppat.1001030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet 2010; 6:e1001191; PMID:21124940; http://dx.doi.org/ 10.1371/journal.pgen.1001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fujino K, Horie M, Honda T, Merriman DK, Tomonaga K. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A 2014; 111:13175-80; PMID:25157155; http://dx.doi.org/ 10.1073/pnas.1407046111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stoye JP. Fv1, the mouse retrovirus resistance gene. Rev Sci Tech 1998; 17:269-77; PMID:9638816 [DOI] [PubMed] [Google Scholar]
- [6].Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 2004; 101:11117-22; PMID:15263098; http://dx.doi.org/ 10.1073/pnas.0402877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Aswad A, Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol Evol 2012; 27:627-36; PMID:22901901; http://dx.doi.org/ 10.1016/j.tree.2012.07.007 [DOI] [PubMed] [Google Scholar]
- [8].Hilditch L, Matadeen R, Goldstone DC, Rosenthal PB, Taylor IA, Stoye JP. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci U S A 2011; 108:5771-6; PMID:21436027; http://dx.doi.org/ 10.1073/pnas.1100118108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kobayashi Y, Horie M, Tomonaga K, Suzuki Y. No evidence for natural selection on endogenous borna-like nucleoprotein elements after the divergence of Old World and New World monkeys. PLoS One 2011; 6:e24403; PMID:21912690; http://dx.doi.org/ 10.1371/journal.pone.0024403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sofuku K, Parrish NF, Honda T, Tomonaga K. Transcription Profiling Demonstrates Epigenetic Control of Non-retroviral RNA Virus-Derived Elements in the Human Genome. Cell Rep 2015; 12:1548-54; PMID:26321645; http://dx.doi.org/ 10.1016/j.celrep.2015.08.007 [DOI] [PubMed] [Google Scholar]
- [11].Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res 2005; 33:2374-83; PMID:15860774; http://dx.doi.org/ 10.1093/nar/gki531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Parrish NF, Fujino K, Shiromoto Y, Iwasaki YW, Ha H, Xing J, Makino A, Kuramochi-Miyagawa S, Nakano T, Siomi H, et al.. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA 2015; 21:1691-703; PMID:26283688; http://dx.doi.org/ 10.1261/rna.052092.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 2012; 26:2361-73; PMID:23124062; http://dx.doi.org/ 10.1101/gad.203786.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 2010; 37:7-19; PMID:20129051; http://dx.doi.org/ 10.1016/j.molcel.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012; 482:331-8; PMID:22337052; http://dx.doi.org/ 10.1038/nature10886 [DOI] [PubMed] [Google Scholar]
- [16].Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science 2006; 313:363-7; PMID:16778019; http://dx.doi.org/ 10.1126/science.1130164 [DOI] [PubMed] [Google Scholar]
- [17].Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006; 442:199-202; PMID:16751776 [DOI] [PubMed] [Google Scholar]
- [18].Hirano T, Iwasaki YW, Lin ZY-C, Imamura M, Seki NM, Sasaki E, Saito K, Okano H, Siomi MC, Siomi H. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 2014; 20:1223-37; PMID:24914035; http://dx.doi.org/ 10.1261/rna.045310.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci U S A 2006; 103:18662-7; PMID:17124176; http://dx.doi.org/ 10.1073/pnas.0605300103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol 2006; 2:e157; PMID:17166054; http://dx.doi.org/ 10.1371/journal.pcbi.0020157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim A, Terzian C, Santamaria P, Pélisson A, Purd'homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci U S A 1994; 91:1285-9; PMID:8108403; http://dx.doi.org/ 10.1073/pnas.91.4.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yamamoto Y, Watanabe T, Hoki Y, Shirane K, Li Y, Ichiiyanagi K, Kuramochi-Miyagawa S, Toyoda A, Fujiyama A, Oginuma M, et al.. Targeted gene silencing in mouse germ cells by insertion of a homologous DNA into a piRNA generating locus. Genome Res 2013; 23:292-9; PMID:23132912; http://dx.doi.org/ 10.1101/gr.137224.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol 1956; 21:197-216; PMID:13433592; http://dx.doi.org/ 10.1101/SQB.1956.021.01.017 [DOI] [PubMed] [Google Scholar]
- [24].Livak KJ. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics 1990; 124:303-16; PMID:1689686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kalmykova AI, Dobritsa AA, Gvozdev VA. Su(Ste) diverged tandem repeats in a Y chromosome of Drosophila melanogaster are transcribed and variously processed. Genetics 1998; 148:243-9; PMID:9475736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW, Duckett CS. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem 2005; 280:22222-32; PMID:15799966; http://dx.doi.org/ 10.1074/jbc.M501928200 [DOI] [PubMed] [Google Scholar]
- [27].Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 2009; 323:393-6; PMID:19150848; http://dx.doi.org/ 10.1126/science.1167375 [DOI] [PubMed] [Google Scholar]
- [28].Shimizu A, Nakatani Y, Nakamura T, Jinno-Oue A, Ishikawa O, Boeke JD, Takeuchi Y, Hoshino H. Characterisation of cytoplasmic DNA complementary to non-retroviral RNA viruses in human cells. Sci Rep 2014; 4:5074; PMID:24875540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu H, Fu Y, Li B, Yu X, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol Biol 2011; 11:276; PMID:21943216; http://dx.doi.org/ 10.1186/1471-2148-11-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, Peng Y, Ghabrial SA, Yi X. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol 2010; 84:11876-87; PMID:20810725; http://dx.doi.org/ 10.1128/JVI.00955-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 2012; 13:283-96; PMID:22421730; http://dx.doi.org/ 10.1038/nrg3199 [DOI] [PubMed] [Google Scholar]
- [32].Zhdanov VM. Integration of viral genomes. Nature 1975; 256:471-3; PMID:51475; http://dx.doi.org/ 10.1038/256471a0 [DOI] [PubMed] [Google Scholar]
- [33].Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh M-C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol 2013; 14:396-403; PMID:23435119; http://dx.doi.org/ 10.1038/ni.2542 [DOI] [PubMed] [Google Scholar]


