Figure 1.
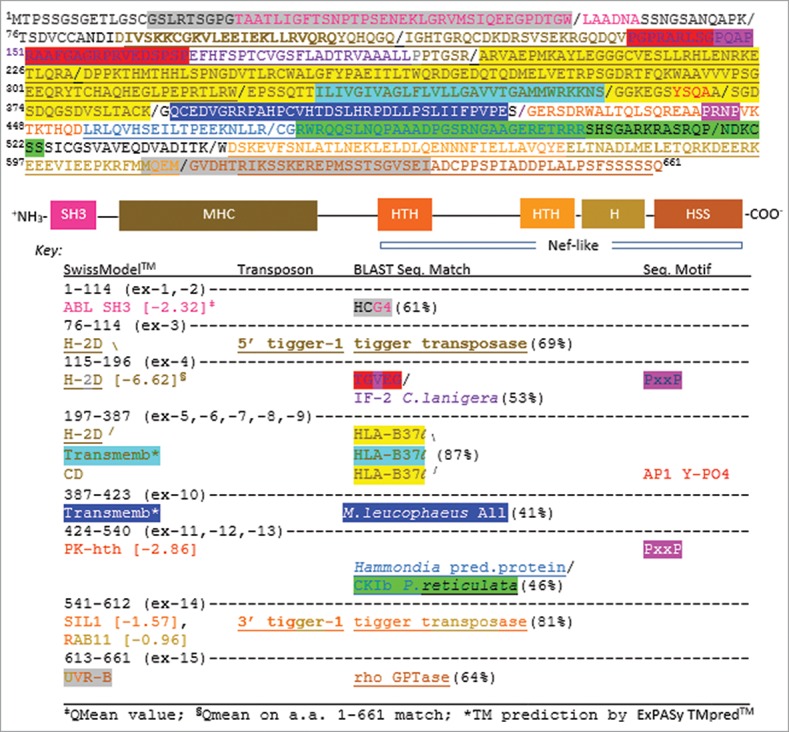
The predicted protein of Loc-103275158 annotated by sequence homology and structural homology. (Top) Amino acid (a.a.) sequence of the orf (exon separation indicated by “/ ” symbols). Color coding matches the (Middle) diagram of the full protein by structural homology to solved domains as determined by SwissModel™ searches on the amino acid sequence shown. (Bottom) The Key shows the locations within the a.a. sequence of the indicated structural domains, the TE footprints of tigger-1, the best sequence matches by BLAST, and motifs associated with known kinases and intracellular trafficking proteins. Gene abbreviations are GenBank annotations (www.ncbi.nlm.nih.gov). H (helix), HTH (helix-turn-helix), HSS (helix-strand-strand).
