Figure 2.
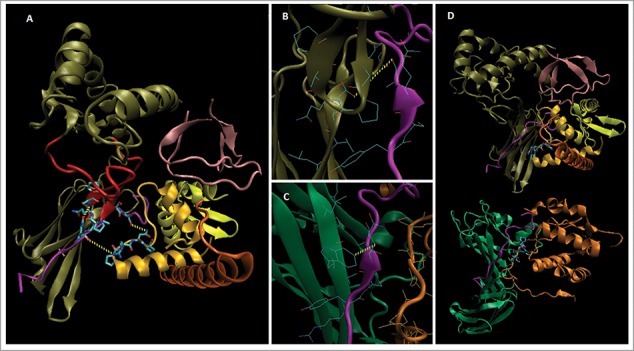
Model structure of Loc-103275158 predicted protein. SwissModel™ (www.expasy.org) was used to generate a homology-based model for 71% of the protein (A). The HLA-A CD from PDB 4EMZ (magenta) was docked at R282 based on homology with the W408:peptide region of 4EMZ (B, C; see text). The Nef-like C-terminal domain was built from matched sub-domain structures in SwissModel™ (see Fig. 1), and docked to the CD peptide on the opposite face from the α3 domain; thus, forming a CD binding groove similar to that of Nef:AP1mu (A-D). As in 4EMZ, a PxxP motif [in the PK-HTH (lt. orange), a.a. P442-P445 (licorice side chains)] was docked to the CD peptide, and the Nef-like structure (A) was built from subdomain homology with Nef (dark orange, D (bottom) after this placement (see D, for overall structural comparison with Nef:MHC-I CD:AP1mu). MHC-I like structure [tan (β-loop in red), CD contacts in licorice]; CD peptide (magenta, Q322 licorice); SH3-like N-terminus (pink); Nef-like C-terminus (lt. orange, yellow, dk. orange, PxxP licorice); AP1mu (green); Nef (dk. orange, PxxP licorice); not shown a.a. in black, including the two transmembrane regions (highlighted lt. blue and blue), Figure. 1 sequence.
