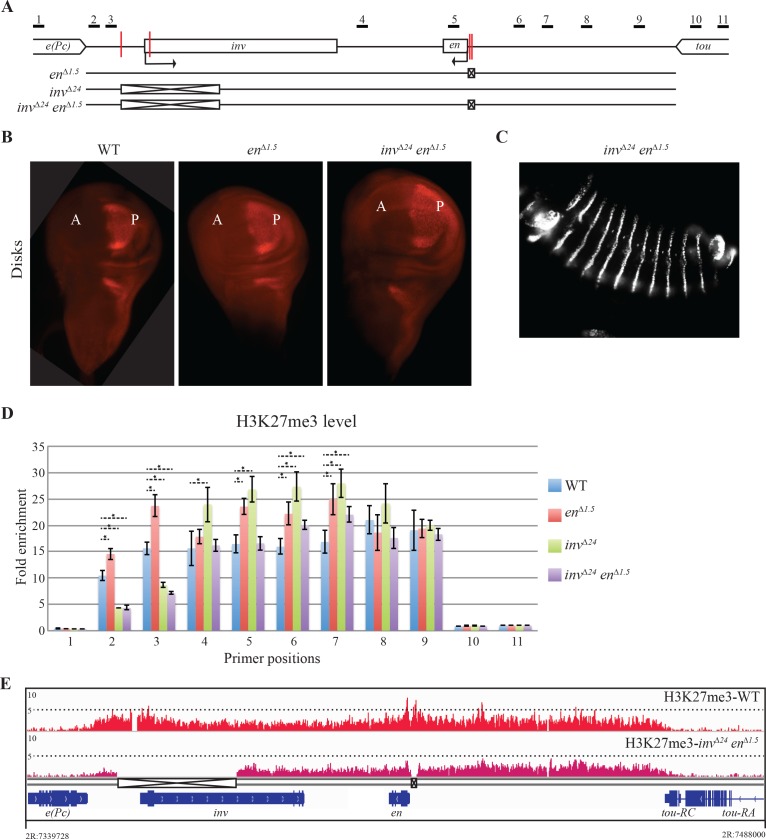
Fig 2. Removal of characterized PREs does not cause mis-expression of en or loss of the H3K27me3 domain.
(A) Line diagram of the inv-en domain showing the extent of deletions in en∆1.5 (2R:7415804..7417265), inv∆24 (2R:7357207..7381178) and inv∆24en∆1.5 mutant flies using crossed boxes, characterized PREs are highlighted with red vertical lines. (B) Immunostaining with anti-En antibody in WT, en∆1.5 and inv∆24en∆1.5 imaginal wing disks. En is expressed in the Posterior compartment (P). PcG proteins repress En expression in the Anterior compartment (A). At least 30 individual wing discs were examined for each genotype. (C) Immunostaining with anti-En antibody in inv∆24en∆1.5 embryo. En is expressed in the stripes, note that there is no en mis-expression observed in between stripes. (D) Quantification of H3K27me3 by ChIP-qPCR over the inv-en domain in larval discs and brains of the genotypes listed. Small horizontal black lines in Fig 2A show regions amplified by qPCR. Results are shown as fold enrichment over background signal and are the average of three independent biological samples with three replicates each (mean±SEM). Statistical analysis of differential H3K27me3 accumulation was performed using Student’s t-test, P-values ≤ 0.05. Only the significant differences with WT are shown with ‘*’. (E) ChIP-seq distribution H3K27me3 in WT and inv∆24en∆1.5 over the inv-en domain is shown.