Figure 1. Topology and evolution.
(A) Topology is concerned with properties of objects that are invariant under continuous deformations. For instance, a “B”-shaped space can be continuously deformed into an “8”-shaped space. Both have one connected piece and two inequivalent loops. These topological invariants are counted by Betti numbers, bn. Similarly, a circumference always has one connected component and a loop, no matter how it is deformed, as long as nothing is cut or pasted.
(B) A prominent tool in algebraic topology are simplicial complexes. These are finite set representations of the original space that share the same topology. Here, we present a simplicial complex that describes the topology of a circumference. The simplicial complex is given in terms of a finite set of elements (3 points and 3 segments). Algebraic operations on the simplicial complex can extract the topological features of the original circumference.
(C) Topological data analysis infers the topological features of a space from a finite set of sampled points by assigning simplicial complexes to the data. One such construction consists of taking balls of fixed radius ε centered on the points. Points at the center of intersecting balls are connected in the simplicial complex. From the resulting complex, it is possible to extract topological features associated to the data at scale ε.
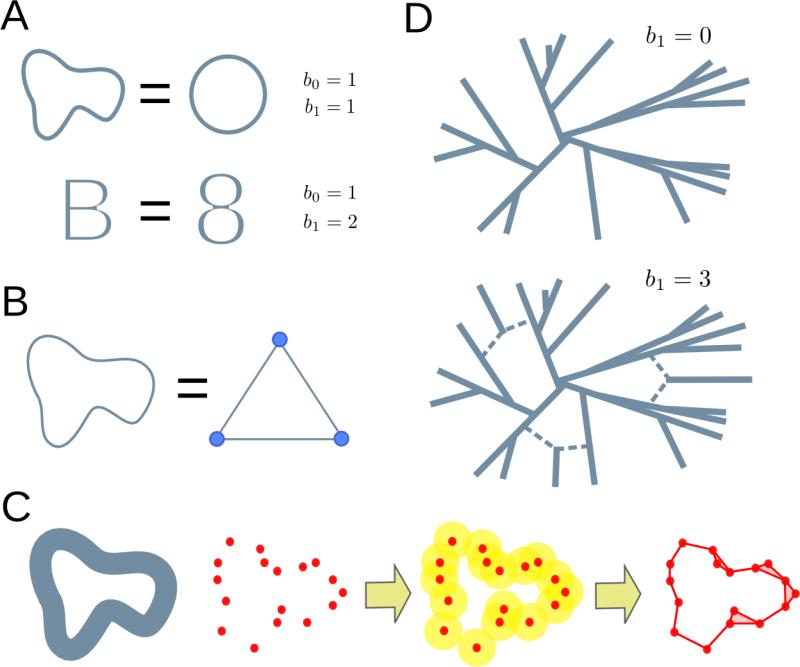
(D) In the context of evolution, genomic sequences can be represented as points in a high dimensional space, where the distance between points is given by the Hamming distance between the corresponding sequences. In the absence of back-mutation and recombination, subsequent mutations can only increase the distance between genomes, and the evolutionary space of the system does not have loops. When recombination events are present, the evolutionary space contains loops, whose presence can be inferred from the finite genomic sample using TDA methods.