Table 2.
Identification, sub-cellular localization, and quantitative analysis of differentially expressed proteins.
| S.N. (U/D) | Accession no. | Name of protein | Mr (da) | pI | M. Score | Location | Relative spot intensities | ||
|---|---|---|---|---|---|---|---|---|---|
| Low N effect | eCO2 effect | eCO2 + Low N effect | |||||||
| 1 (U) | gi|111608879 | α-1,4-glucan phosphorylase | 8852 | 5.17 | 134 | Chloroplast | 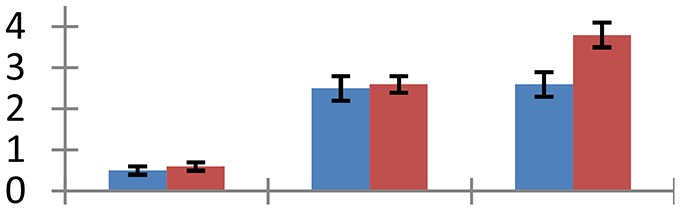 |
||
| 2 (U) | gi|226533870 | cp31BHv | 18,293 | 4.85 | 86 | Nucleus, chloroplast | 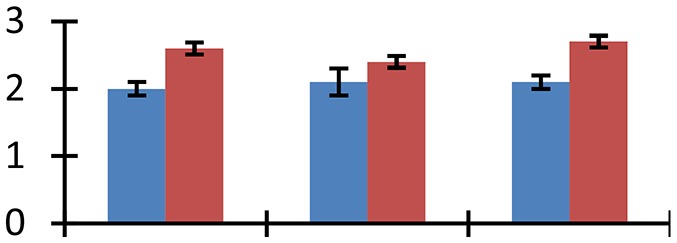 |
||
| 3 (U) | gi|145327751 | GLYOXYLASE I 7 | 15,564 | 5.22 | 74 | Peroxisome | 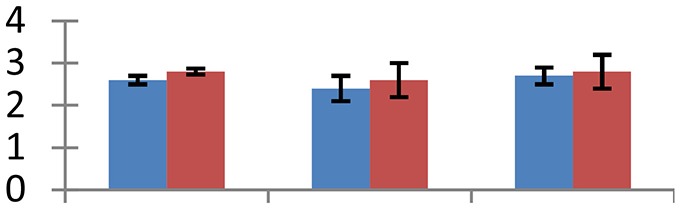 |
||
| 4 (U) | gi|88909669 | L-ascorbate peroxidase 5 | 33,852 | 5.84 | 94 | Chloroplast | 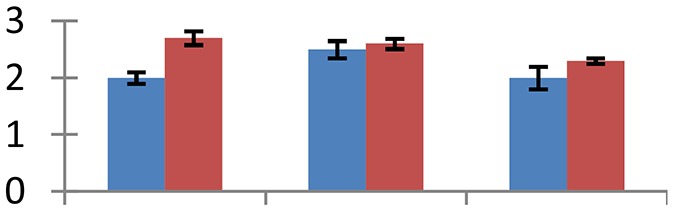 |
||
| 5 (D) | gi|1174745 | Triosephosphate isomerase, chloroplastic | 31,955 | 6.10 | 148 | Chloroplast, cytosol | 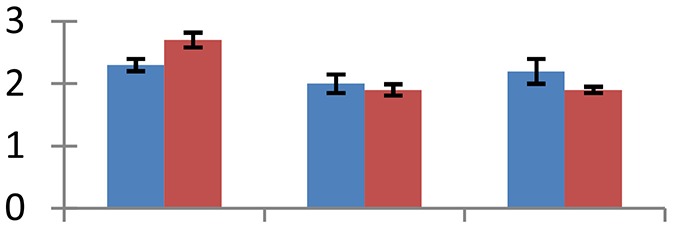 |
||
| 6 (U) | gi|259490070 | Lipid binding protein precursor | 12,904 | 6.69 | 59 | Membrane | 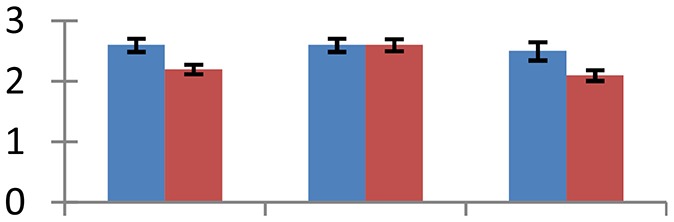 |
||
| 7 (U) | gi|6523032 | Flavonol synthase-like protein | 36,401 | 5.33 | 94 | Cytoplasm, nucleus | 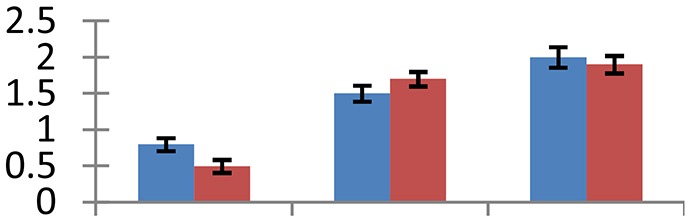 |
||
| 8 (U) | gi|15229664 | MYB77; DNA binding/transcription factor | 34,318 | 6.07 | 53 | Nucleus | 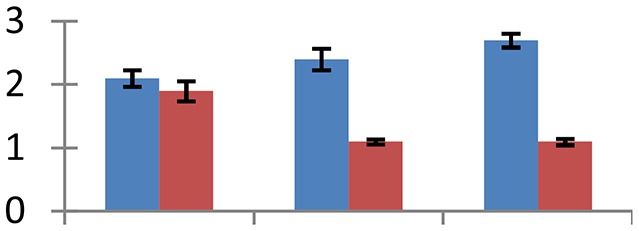 |
||
| 9 (D) | gi|521301355 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 53,901 | 6.16 | 351 | Chloroplast | 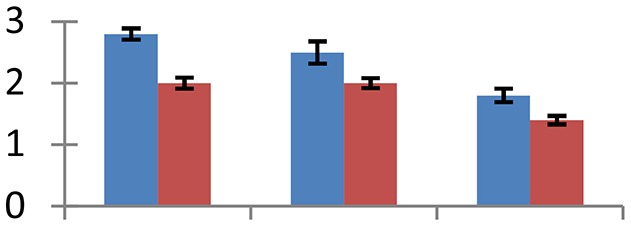 |
||
| 10 (D) | gi|15220615 | Chlorophyll a-b binding protein 1 | 24,836 | 5.11 | 112 | Chloroplast | 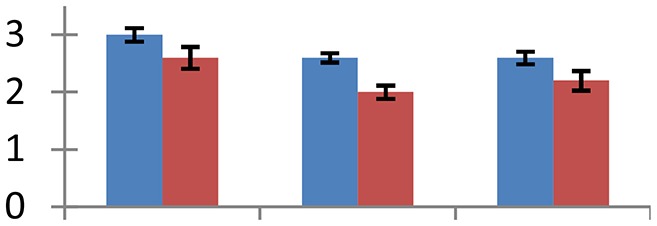 |
||
| 11 (D) | gi|226493349 | 3-isopropylmalate dehydratase large subunit 2 | 53,901 | 7.05 | 147 | Chloroplast | 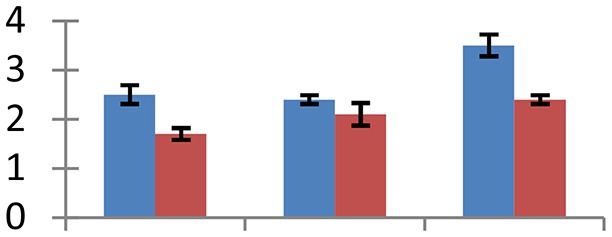 |
||
| 12 (U) | gi|162135976 | Sterolesin-B | 28,873 | 5.63 | 55 | 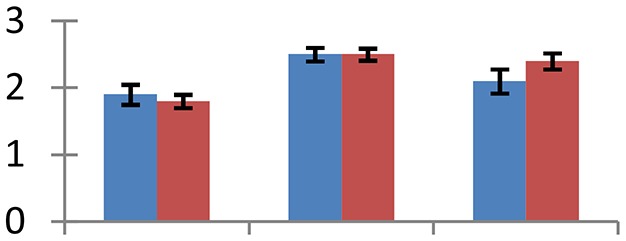 |
|||
| 13 (U) | gi|37653227 | PII-like protein | 25,729 | 9.84 | 160 | Chloroplast | 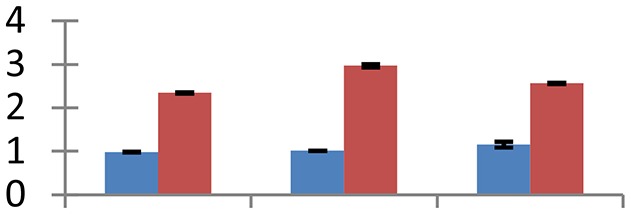 |
||
| 14 (U) | gi|21322655 | ADP glucose pyrophosphatase | 21,172 | 5.62 | 83 | Chloroplast | 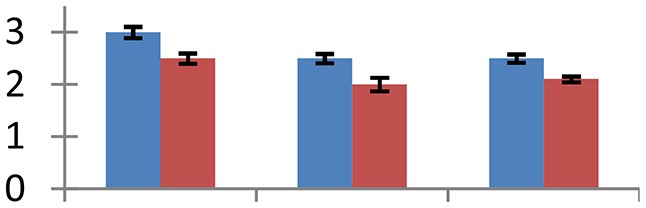 |
||
| 15 (U) | gi|334183671 | Glutamine synthetase | 46,852 | 5.96 | 120 | Chloroplast/mitochondria | 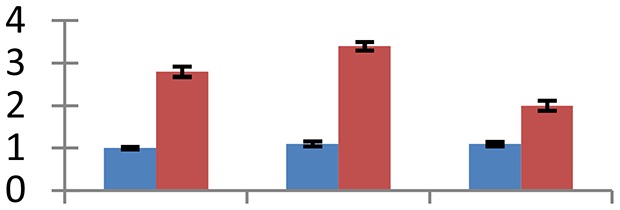 |
||
| 16 (U) | gi|255073229 | Glutathione transferase | 25,098 | 6.35 | 76 | Cytosol | 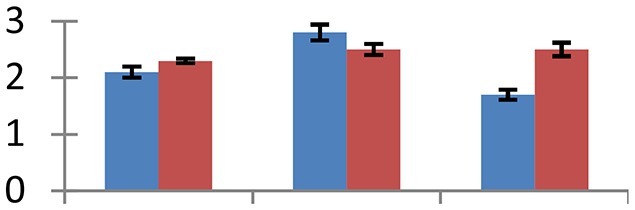 |
||
| 17 (U) | gi|297382831 | Beta-amylase | 24,276 | 5.59 | 64 | Chloroplast, vacuole, cytosol | 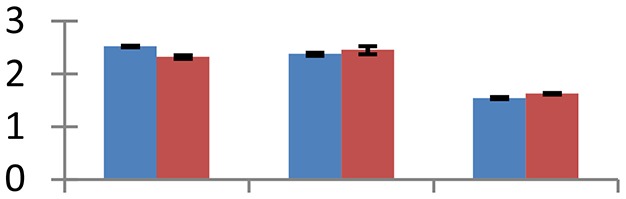 |
||
| 18 (D) | gi|27735224 | Ribulose bisphosphate carboxylase small chain 2B | 20,801 | 6.29 | 232 | Chloroplast | 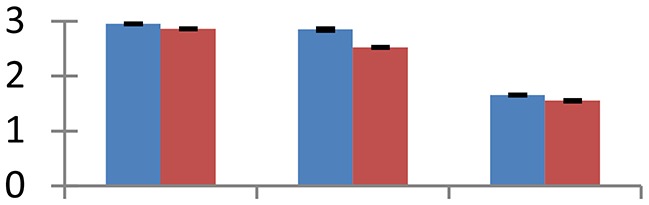 |
||
| 19 (U) | gi|224015956 | RADIALIS | 9381 | 8.29 | 32 | Nucleus | 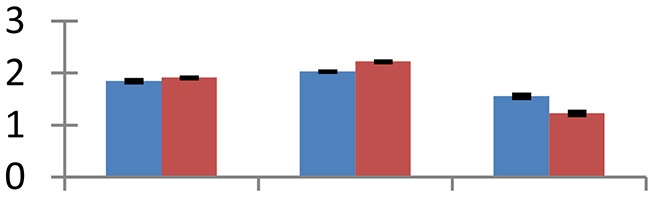 |
||
| 20 (U) | gi|31415978 | Putative TNP2-like transposase | 116,347 | 7.91 | 60 | Nucleus | 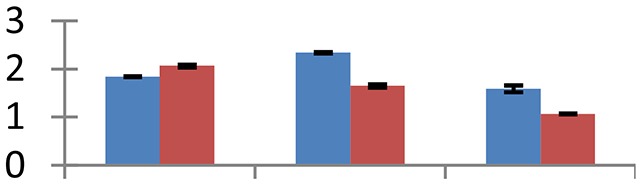 |
||
| 21 (D) | gi|108706066 | Cytoplasmic aconitate hydratase | 99,314 | 5.73 | 151 | Cytosol | 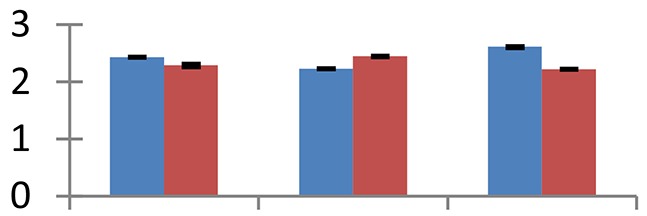 |
||
| 22 (U) | gi|159157513 | Putative chaperonin 60 beta precursor | 64,721 | 5.68 | 158 | Mitochondria | 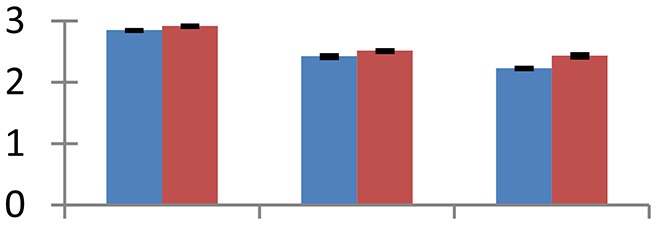 |
||
| 23 (U) | gi|2244775 | Salt-inducible protein homolog | 65,905 | 5.96 | 51 | Nucleus | 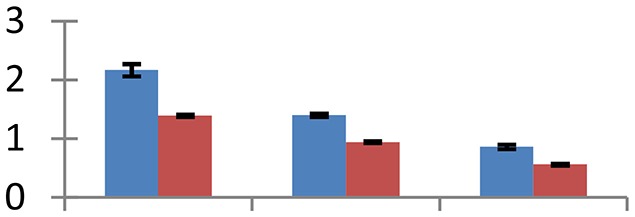 |
||
| 24 (U) | gi|15231865 | AGD6; ARF GTPase activator/DNA binding/zinc ion binding | 49,222 | 6.57 | 99 | Golgi apparatus | 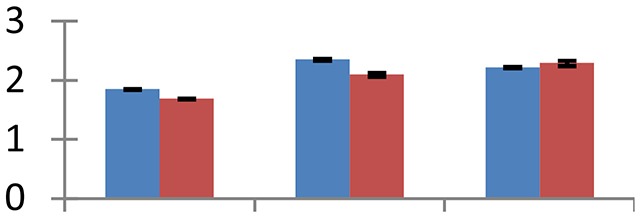 |
||
| 25 (D) | gi|49574618 | Ribosomal protein L14 | 13,216 | 9.71 | 54 | Ribosome | 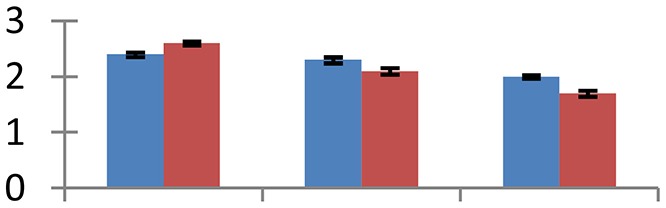 |
||
| 26 (D) | gi|114329695 | Ribosomal protein S19 | 10,689 | 10.90 | 50 | Ribosome, chloroplast, mitochondria | 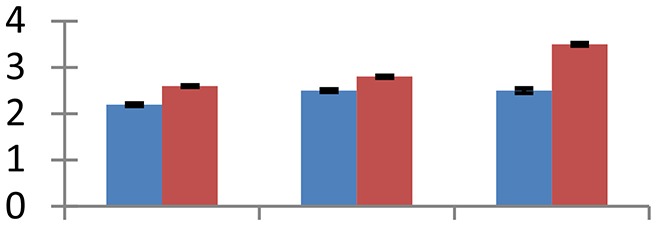 |
||
| 27 (U) | gi|325947633 | Salt overly sensitive 2 | 20,331 | 4.94 | 33 | Plasma membrane | 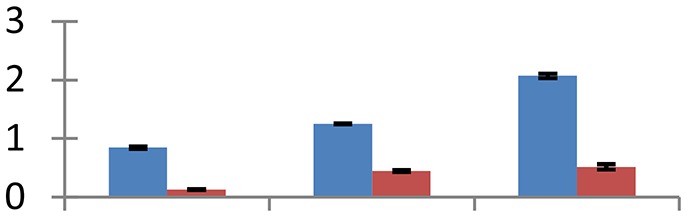 |
||
| 28 (U) | gi|100831 | Photosystem II oxygen-evolving complex protein 1 | 33,596 | 8.13 | 160 | Chloroplast | 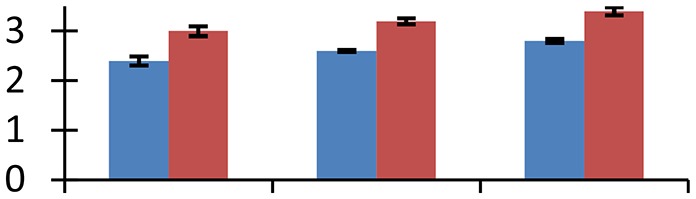 |
||
| 29 (U) | gi|1263097 | MYB-related protein | 33,799 | 6.07 | 133 | Nucleus | 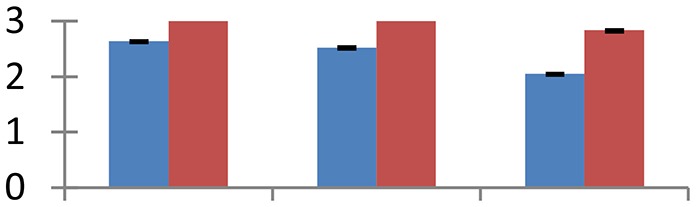 |
||
| 30 (D) | gi|2546952 | Translation elongation factor-TU | 27,375 | 5.09 | 99 | Plastid, mitochondria, cytosol | 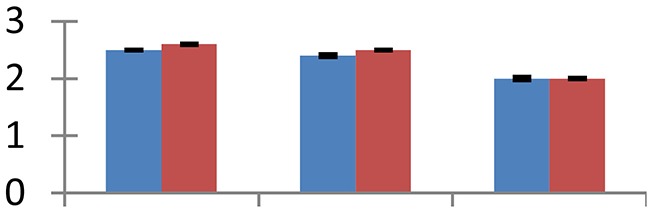 |
||
| 31 (U) | gi|4090259 | Ubiquitin-conjugating enzyme E2 | 18,891 | 7.01 | 75 | Nucleus, cytosol | 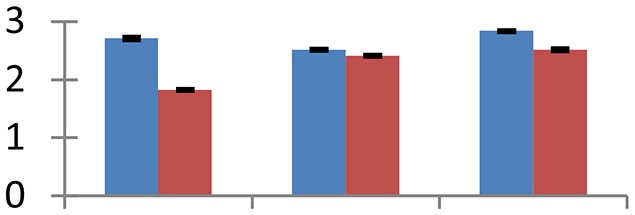 |
||
| 32 (U) | gi|145666464 | Protein disulfide isomerase | 56,921 | 5.01 | 149 | ER | 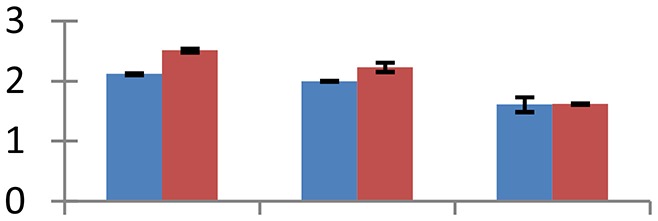 |
||
| 33 (U) | gi|12643259 | Rubisco activase chloroplast precursor | 51,886 | 5.43 | 138 | Chloroplast | 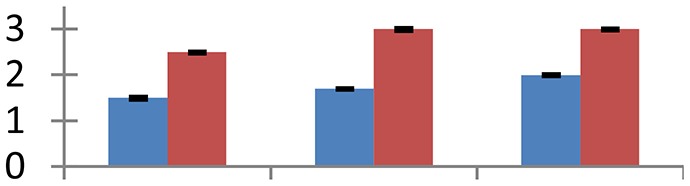 |
||
| 34 (U) | gi|330255651 | 3-Ketoacyl-CoA synthase | 46,810 | 9.02 | 65 | Plasma membrane | 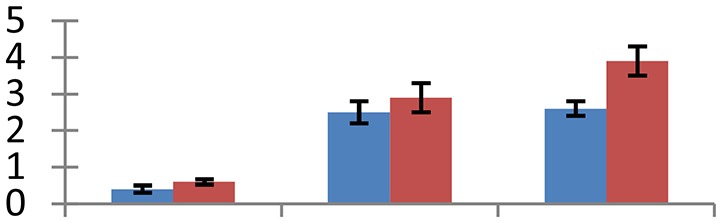 |
||
| 35 (U) | gi|297807473 | GDSL-motif lipase/hydrolase family protein | 43,124 | 8.76 | 34 | chloroplast stroma | 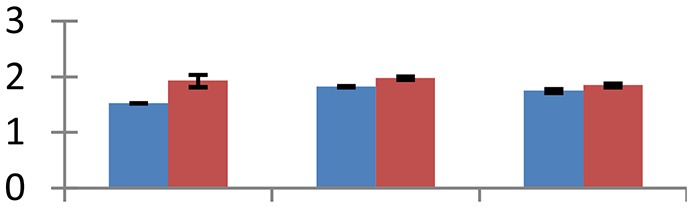 |
||
| 36 (U) | gi|1572627 | Cu/Zn superoxide dismutase | 20,312 | 5.35 | 130 | Cytosol | 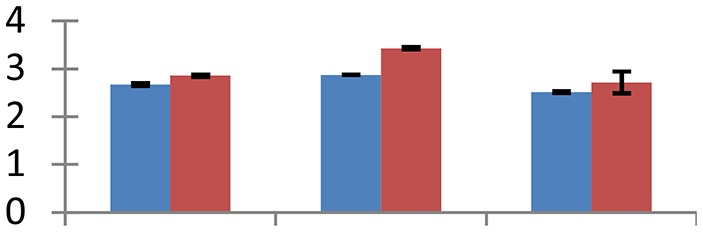 |
||
| 37 (U) | gi|15231902 | Phosphatidylinositol-4-phosphate 5-kinase family protein | 82,129 | 8.63 | 133 | Plasma membrane | 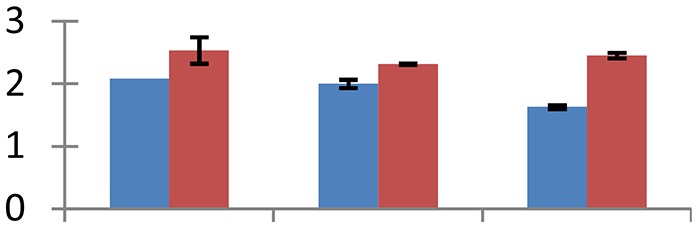 |
||
| 38 (U) | gi|30686609 | PUB23 (PLANT U-BOX 23); ubiquitin-protein ligase | 46,648 | 8.28 | 92 | Cytosol | 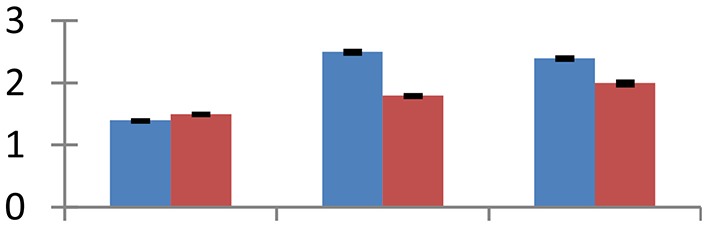 |
||
| 39 (D) | gi| 15229231 | Glyceraldehyde 3-phosphate dehydrogenase | 37,366 | 6.62 | 141 | Plastid, mitochondria | 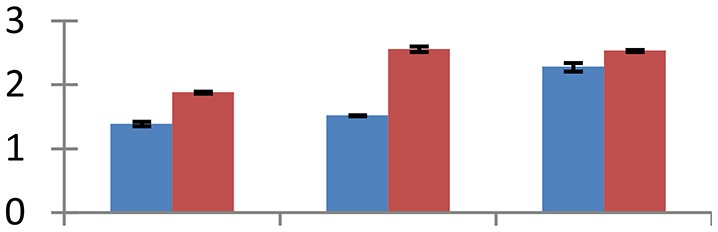 |
||
| 40 (U) | gi|729477 | RecName: Ferredoxin-NADP reductase | 41,622 | 8.54 | 139 | Chloroplast stroma, thylakoid membrane | 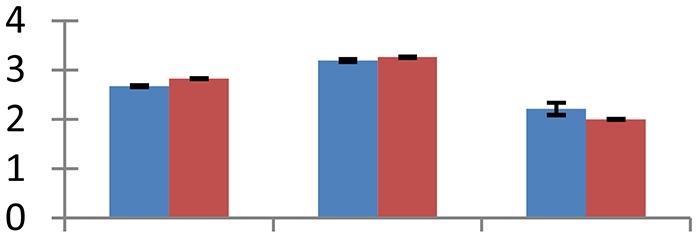 |
||
| 41 (D) | gi|296932709 | Granule-bound starch synthase I | 13,696 | 9.76 | 51 | Chloroplast | 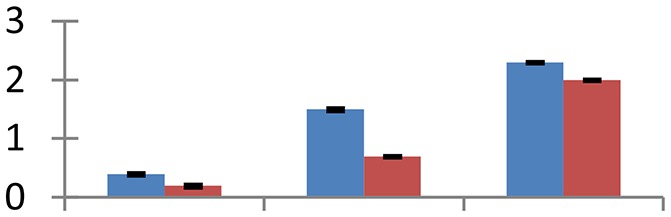 |
||
| 42 (U) | gi|50252278 | Flavin-containing monooxygenase | 42,795 | 8.75 | 65 | ER | 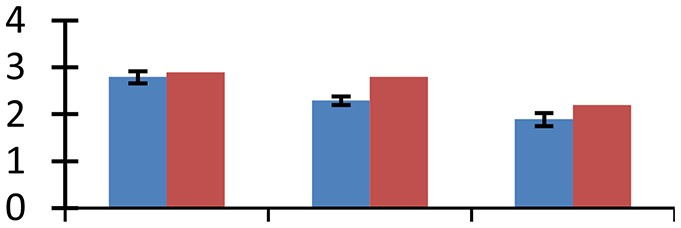 |
||
| 43 (D) | gi|186886309 | ATPase F1 alpha subunit | 38,233 | 8.27 | 134 | Mitochondria | 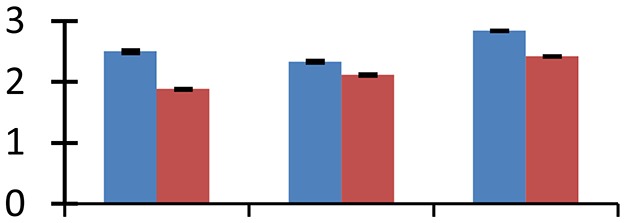 |
||
| 44 (D) | gi|260780700 | Acetyl-CoA carboxylase beta subunit | 40,007 | 9.02 | 53 | Cytoplasm, plastid | 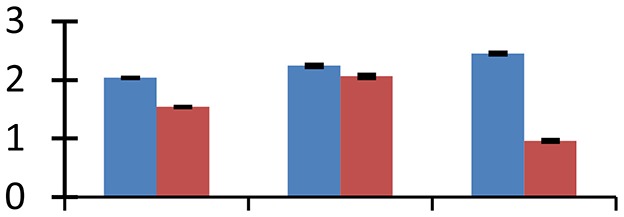 |
||
| 45 (U) | gi|2570499 | 23 kDa polypeptide of photosystem II | 27,772 | 9.06 | 94 | Chloroplast | 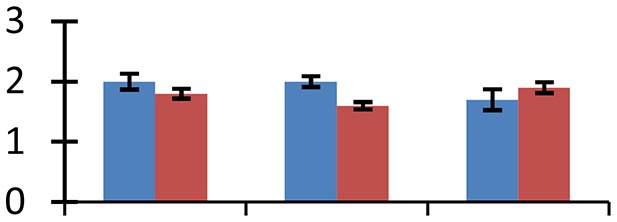 |
||
| 46 (U) | gi|83700338 | Chloroplast-localized cyclophilin | 16,127 | 8.48 | 146 | Chloroplast | 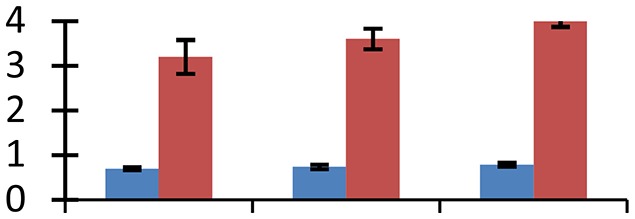 |
||
| 47 (D) | gi|52353463 | ACC oxidase | 36,852 | 4.97 | 134 | Cytosol | 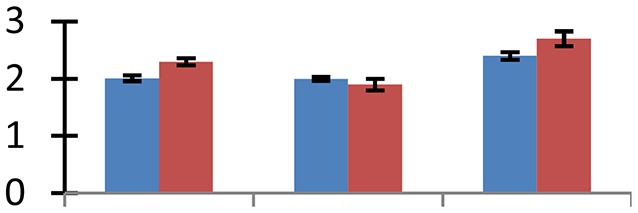 |
||
| 48 (D) | gi|217075182 | Unknown | 19,372 | 8.81 | 39 | 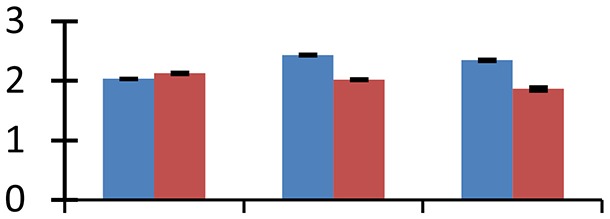 |
|||
| 49 (U) | gi|145329965 | TTN7 (TITAN7); ATP binding/protein binding | 138,856 | 6.05 | 144 | Nucleus | 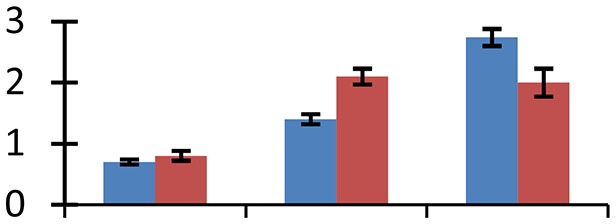 |
||
| 50 (D) | gi|56785387 | Hypothetical protein [Oryza sativa Japonica Group] | 8515 | 11.31 | 40 | 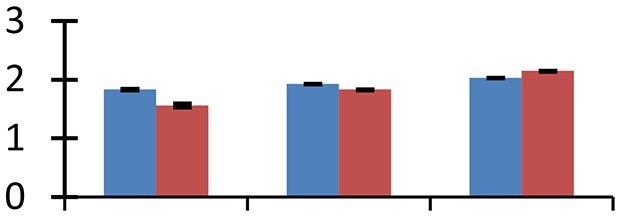 |
|||
| 51 (U) | gi|21537409 | Osmotin-like protein | 27,148 | 6.34 | 135 | Cell-wall | 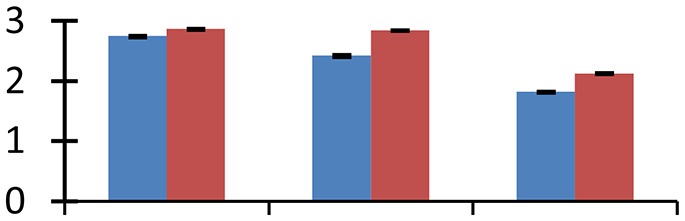 |
||
| 52 (D) | gi|22296371 | Putative 29 kDa ribonucleoprotein A, chloroplast precursor | 28,126 | 4.75 | 165 | Nucleus | 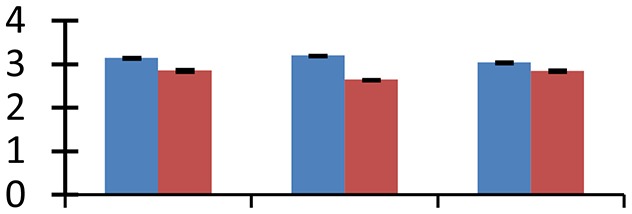 |
||
| 53 (D) | gi|41584495 | SMC3 | 139,879 | 6.07 | 54 | Cytoplasm, nucleus | 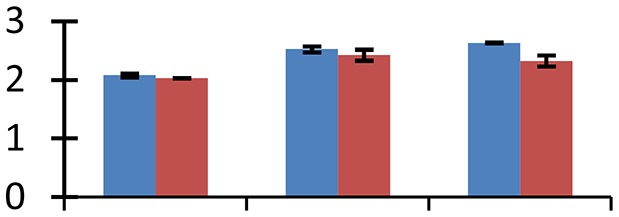 |
||
| 54 (D) | gi|79595267 | ATPase | 209,868 | 6.50 | 169 | Plasma membrane, tonoplast | 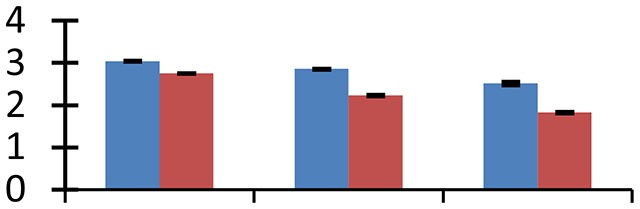 |
||
| 55 (U) | gi|255575181 | Carnitine racemase | 27,262 | 7.64 | 32 | Nucleus, peroxisome | 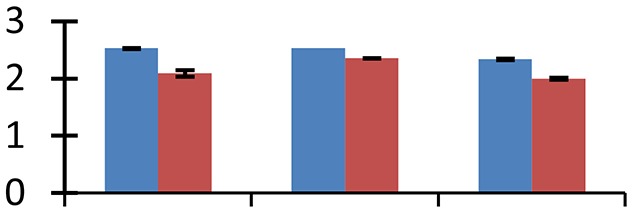 |
||
| 56 (U) | gi|297807151 | Hypothetical protein ARALYDRAFT_909075 | 27,949 | 5.57 | 45 | 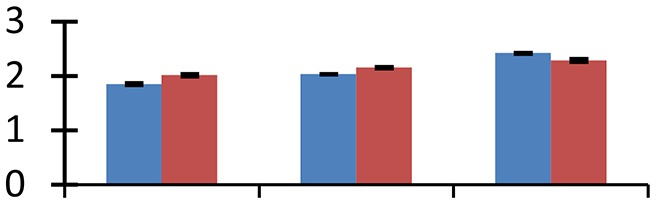 |
|||
| 57 (D) | gi|260619528 | LIM1 | 21,920 | 9.07 | 124 | Nucleus, cytosol | 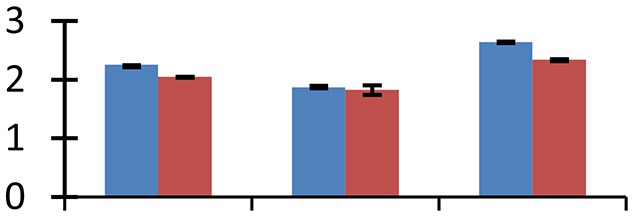 |
||
| 58 (D) | gi|69216892 | RNA polymerase beta chain | 79,308 | 9.10 | 217 | Nucleus | 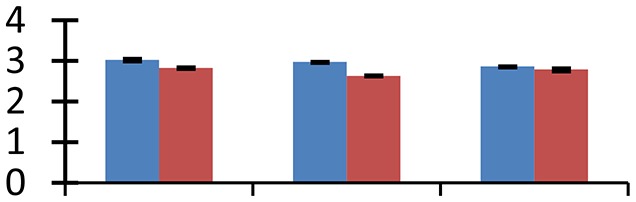 |
||
| 59 (U) | gi|21397266 | Small heat shock protein | 25,122 | 6.26 | 61 | Nucleus | 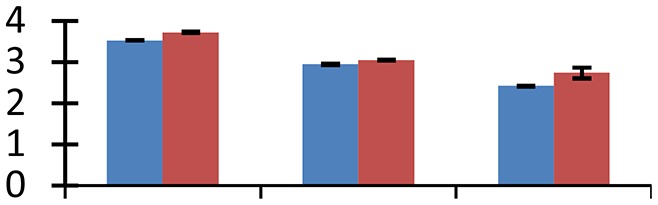 |
||
| 60 (U) | gi|269969449 | Iron deficiency-specific protein | 374,514 | 5.71 | 78 | Cytosol | 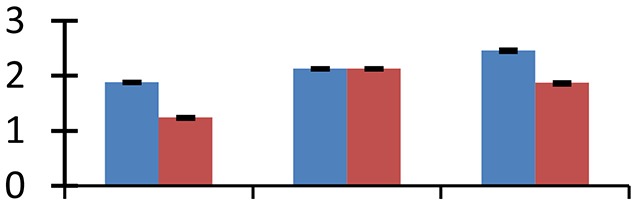 |
||
| 61 (U) | gi|38344419 | OSJNBb0080H08.11 | 202,531 | 8.71 | 46 | 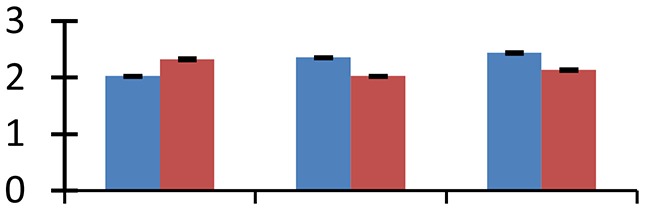 |
|||
| 62 (U) | gi|304571953 | Anthranilic acid methyltransferase 3 | 43,177 | 6.29 | 142 | Cytosol | 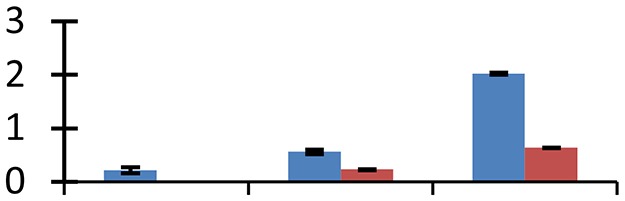 |
||
| 63 (D) | gi|4261643 | Sedoheptulose-1,7-bisphosphatase | 42,068 | 6.26 | 153 | Chloroplast | 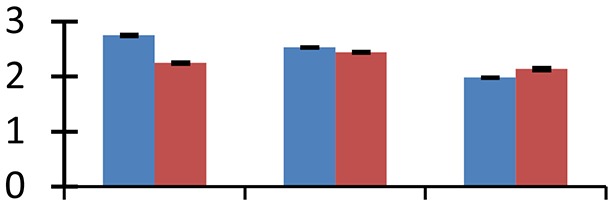 |
||
| 64 (U) | gi|18424254 | Zinc finger (C3HC4-type RING finger) family protein | 44,981 | 6.36 | 88 | Nucleus | 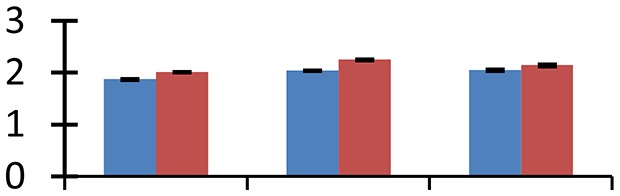 |
||
| 65 (U) | gi|159487407 | NAD-dependent epimerase/dehydratase | 36,518 | 7.13 | 135 | Cytosol | 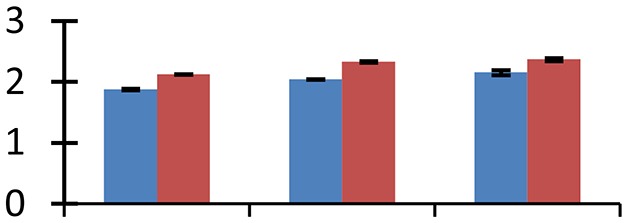 |
||
| 66 (D) | gi|297806809 | Galactosyl transferase GMA12/MNN10 family protein | 52,488 | 7.52 | 95 | Golgi apparatus | 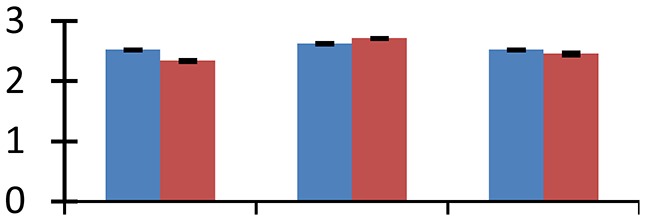 |
||
| 67 (U) | gi|113205313 | “Chromo” domain containing protein | 93,199 | 9.21 | 143 | Thylakoid membrane | 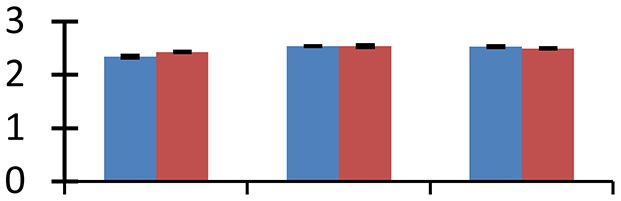 |
||
| 68 (U) | gi|15213029 | Maturase K | 14,517 | 9.86 | 55 | Mitochondria, chloroplast, nucleus | 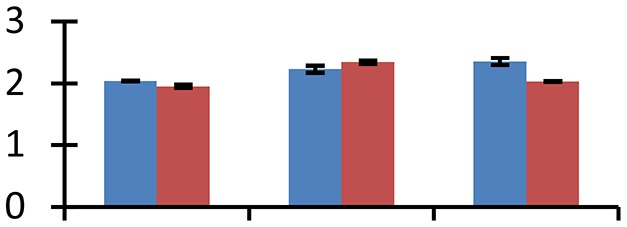 |
||
| 69 (U) | gi|308804756 | Stress-induced protein sti1-like protein (ISS) | 69,580 | 6.03 | 72 | Cytosol, chloroplast | 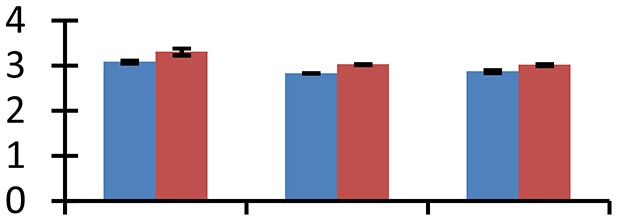 |
||
| 70 (U) | gi|238005590 | Unknown | 9609 | 8.88 | 52 | 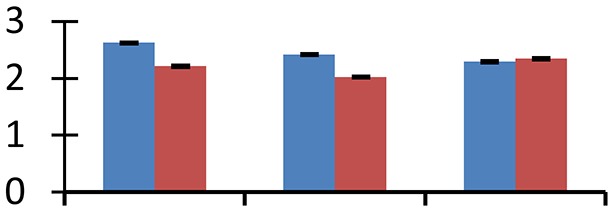 |
|||
| 71 (D) | gi|75180109 | Heme oxygenase 4 | 32,115 | 6.89 | 62 | Chloroplast | 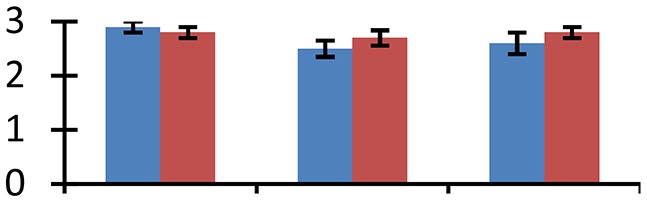 |
||
| 72 (U) | gi|300669644 | Ferredoxin-dependent glutamate synthase 2 | 27,542 | 6.24 | 102 | Chloroplast | 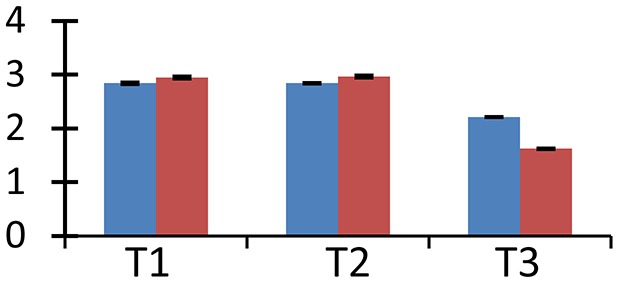 |
||
S.N. (U/D), Spot Number; U, upregulated; D, downregulated; Mr, Molecular Weight; pI, Isoelectric Point; M.Sco, MASCOT Score; eCO2, elevated CO2 level. Y-axis denoted relative spot intensities; X-axis denotes treatments. Red bars indicate Pusa Bold cultivar, and blue bars indicate Pusa Jai Kisan cultivar. Spot volumes were analyzed by PD Quest software. The fold-change of up-regulated protein spot volumes was calculated by treatment (T1-T3)/control (T0), whereas the change fold of down-regulated protein spot volumes was calculated by control (T0)/treatment (T1-T3). T0, ambient CO2 level and sufficient-N; T1, ambient CO2 level and low-N; T2, elevated CO2 level and sufficient-N; T3, low-N and elevated CO2 level. Values are presented as mean of three replicates ± SE.
