FIGURE 10.
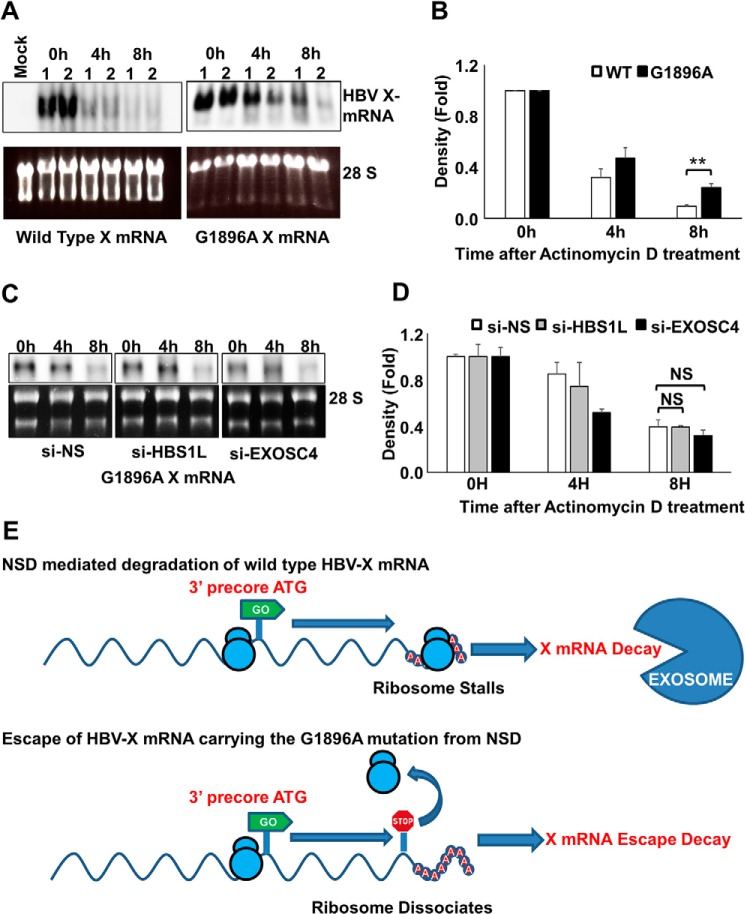
Precore premature stop codon increases HBV X-mRNA stability. A, HepG2 cells were transfected with pcDNA3.1-X or pcDNA3.1-X (G1896A), a construct harboring a stop codon in-frame with the 3′ precore ATG. 48 h later the cells were treated with actinomycin D (8 μg/ml). Total cellular RNA was collected at the indicated time points, and detection of HBV X-mRNA was performed by Northern blot analysis (upper panels). As a loading control, the 28S and 18S ribosomal RNAs in the input samples were stained with ethidium bromide (lower panels). Northern blot data from two independent experiments (1 and 2) were shown. B, HBV X-mRNA density was calculated by Image J software, plotted in the lower panel as -fold difference compared with the density at 0 h. Data from three independent experiments are presented as the mean ± S.D. C, HepG2 cells were transfected with si-NS, si-HBS1L, or si-EXOSC4 as indicated. 24 h later, cells were transfected with pcDNA3.1-X (G1896A). After 48 h, the cells were treated with actinomycin D (8 μg/ml). Total cellular RNA was collected at the indicated time points, and HBV X mRNA was detected by Northern blot analysis (upper panels). As a loading control, the 28S and 18S ribosomal rRNAs in the input samples were visualized (after electrophoresis) by staining with ethidium bromide and imaging under UV illumination (lower panels). D, HBV X-mRNA bands density shown in C were calculated by Image J software, plotted as -fold difference compared with the density at 0 h. Data from three independent experiments are presented as the mean ± S.D. NS, not significant. E, a diagram explaining the possible effect of G1896A mutation leading to the escape of HBV X mRNA from NSD RNA quality control. Statistical significance was measured was measured using two-tailed Student's t test. ** = p < 0.01