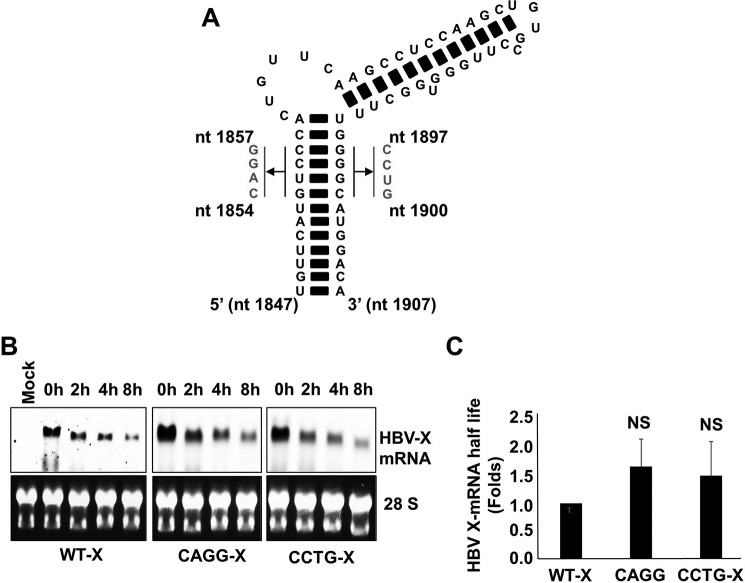
FIGURE 8.
A, schematic showing the mutations introduced to disrupt HBV-epsilon's secondary structure. Either the sequence from nt 1854 to 1857 on the 5′ strand was mutated to 5′-CAGG-3′ or the sequence from nt 1897 to1900 on the 3′ strand was mutated to 5′-CCUG-3′ (nucleotide numbering is based on the HBV-D_IND60 sequence. GenBankTM accession number AB246347.1). B, HepG2 cells were transfected with pcDNA3.1-X, pcDNA3.1-X (5′-CAGG-3′), or pcDNA3.1-X (5′-CCTG-3′). 48 h later the cells were treated with actinomycin D (8 μg/ml). Total cellular RNA was collected at the indicated time points, and HBV X-mRNA levels were determined by Northern blot analysis (upper panels). As a loading control, the 28S and 18S ribosomal RNAs in the input samples were stained with ethidium bromide (lower panels). C, the density of the different HBV X-mRNA bands detected by the Northern blot analysis shown in A were quantified and used to calculate the half-lives of HBV X-mRNA. The half-lives of HBV X-mRNA encoded by the different constructs were plotted as the -fold difference compared with that of wild-type HBV X-mRNA. Data from three experiments are presented as the mean ± S.D. NS, not significant.