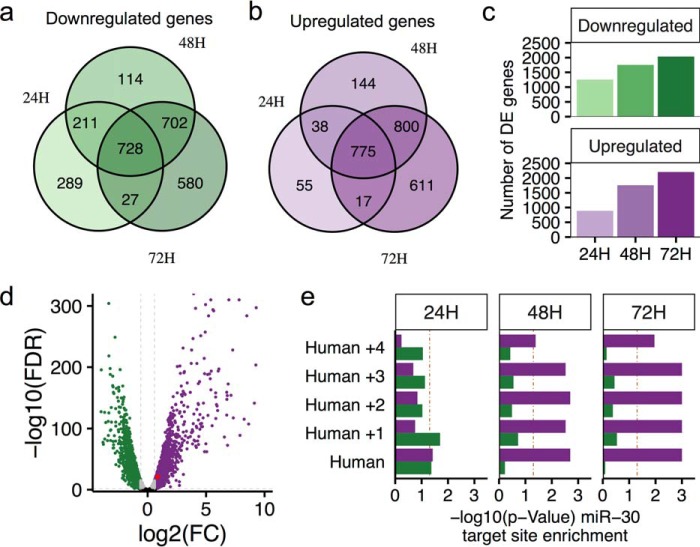
FIGURE 4.
LNA30bcd-treated HIECs undergo robust changes in gene expression over a 3-day time course. a, Venn diagram showing significantly down-regulated genes (fold change or FC < −1.5, FDR < 0.05) across time points in HIECs after 100 nm LNA30bcd transfection compared with mock transfection. b, Venn diagram showing significantly up-regulated genes (FC > 1.5, FDR < 0.05) across time points in HIECs after 100 nm LNA30bcd transfection compared with mock transfection. c, bar graph showing the number of significantly down-regulated and up-regulated genes across time points. d, volcano plot showing differentially expressed genes at 72 h. The red dot shows SOX9. The horizontal dashed line shows p = 0.05, and vertical dashed lines indicate FC = −1.5 and 1.5. e, results of miRhub analysis to test for enrichment of predicted miR-30 target sites in significantly up-regulated (purple) and down-regulated (green) genes at each time point. Our analysis was human-centric. Each row indicates the conservation of the miRNA target site on the gene list, with Human indicating a site found in human genes and Human +1 indicating a site found in human genes and conserved in one additional species, and so on. The vertical dashed line (red) indicates empirical p = 0.05.