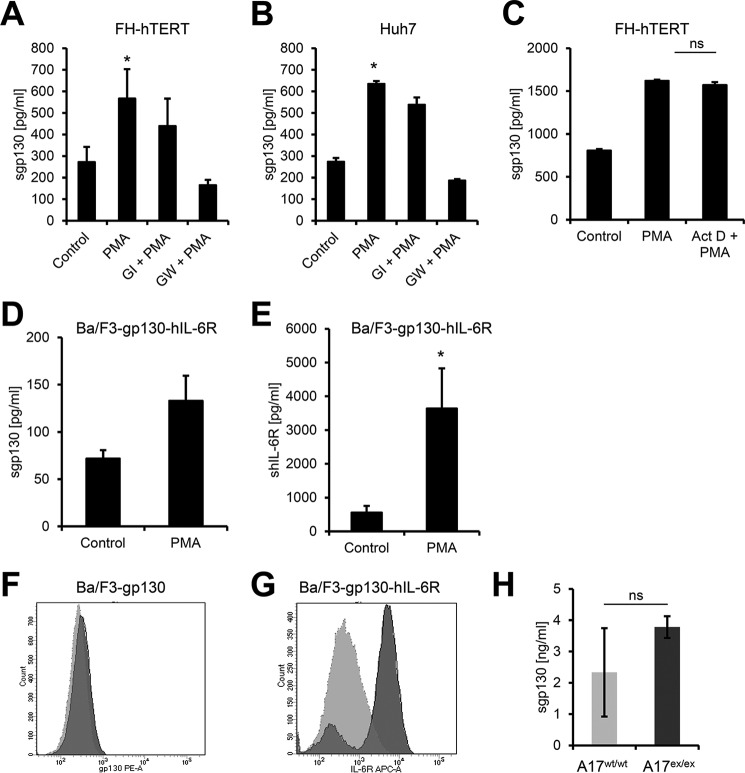
FIGURE 1.
The protease ADAM17 sheds membrane-bound gp130 with low efficiency. A and B, FH-hTERT (A) and Huh7 (B) cells were stimulated with 100 nm PMA for 2 h or treated with DMSO as a control. Cells were pretreated with 3 μm GI (a specific ADAM10 inhibitor) or 3 μm GW (a specific combined ADAM10/ADAM17 inhibitor) 30 min prior to PMA stimulation as indicated. The amount of sgp130 in the cell culture supernatant was analyzed by ELISA. C, FH-hTERT cells were either stimulated with 100 nm PMA for 2 h or treated with DMSO as a control. Where indicated, cells were pretreated with 1 μg/ml actinomycin D (Act D) 30 min prior to stimulation with PMA. The amount of sgp130 in the cell culture supernatant was analyzed by ELISA. D and E, Ba/F3-gp130-hIL-6R cells were stimulated with 100 nm PMA for 2 h or treated with DMSO as a control. The amount of sgp130 (D) or shIL-6R (E) in the cell culture supernatant was analyzed via specific ELISAs. F, analysis of proteolytic cleavage of gp130 in DMSO- (dark gray) or PMA-treated (light gray) Ba/F3-gp130 cells by flow cytometry. G, analysis of proteolytic cleavage of IL-6R in DMSO- (dark gray) or PMA-treated (light gray) Ba/F3-gp130-IL6R cells by flow cytometry. H, analysis of endogenous sgp130 in wild-type or ADAM17ex/ex mice (n = 3 each) by murine sgp130-ELISA. All ELISA data shown are the mean ± S.E. derived from three independent experiments. Flow cytometry data show one representative experiment of three independent experiments with similar outcomes. *, p < 0.05; ns, not significant.