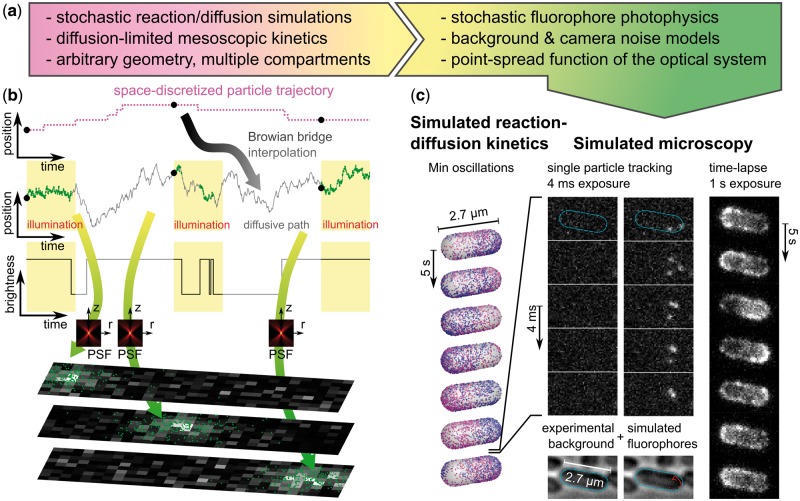
Fig. 1.
Simulated microscopy with SMeagol. (a) Workflow from stochastic reaction–diffusion simulations to images. (b) The microscopy simulation starts from trajectories generated by stochastic reaction–diffusion simulations, fills in stochastic motion and photon emission events between the trajectory points, and finally combines PSF and camera noise models to simulate realistic images. (c) Simulated microscopy of fluorescently labeled MinE proteins in the Min oscillatory system. Left: Stochastic reaction–diffusion simulation. Mid columns: Simulated SPT microscopy using an actual experimental background noise movie with continuous illumination and 4 ms/frame. Right: A simulation of continuous illumination and 1 s/frame renders a conventional (non-single molecule) fluorescence microscopy time-lapse movie. See also Supplementary movies S1, S2 and the Supplementary material for further details (Color version of this figure is available at Bioinformatics online.)