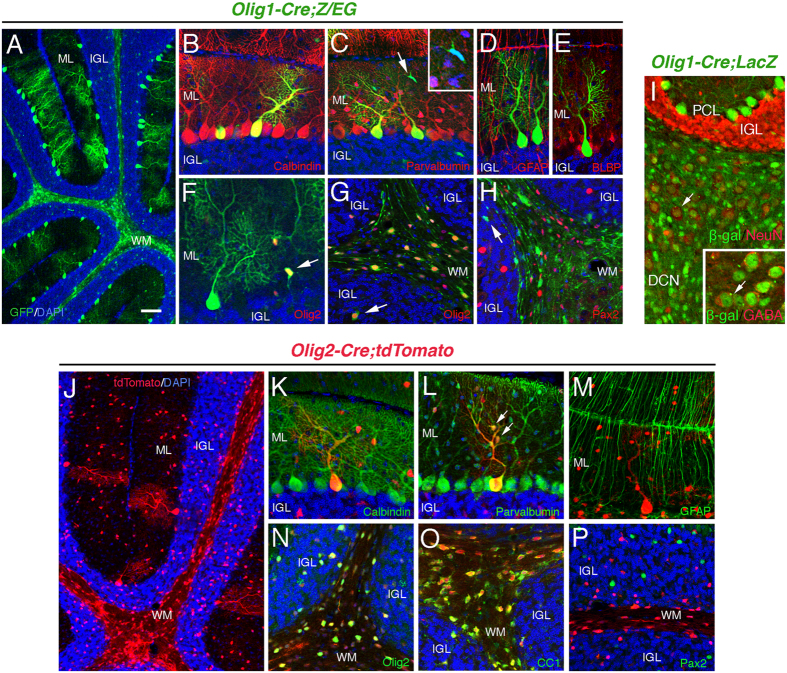
Figure 6. Olig gene-expressing progenitors give rise to PCs and DCN neurons but rarely Pax2+ interneurons in the postnatal cerebellum as revealed by long-term lineage tracing.
Olig1-Cre lineage tracing analysis is performed in the cerebellum of either Z/EG (A–H) reporter line at P18 or ROSA26;LacZ (I) reporter line at P16. Labeled cells with different morphology in the cerebellum of Olig1-Cre;Z/EG mice are revealed by their GFP expression (A). The identity of GFP+ cells are examined by co-staining with cell-type specific markers Calbindin (B), Parvalbumin (C), GFAP (D), BLBP (E), Olig2 (F,G) and Pax2 (H). Some GFP+ cells are indicated as either negative (arrows in C and H) or positive (arrows in F,G) for marker staining. The high magnification insert in (C) confirms the cellular identity of the indicated GFP+ cell by showing the presence of DAPI+ nucleus. Many labeled cells in the cerebellar DCN of Olig1-Cre;LacZ reporter mice are observed by anti-β-gal staining, some of which are NeuN+ (arrow, I) and GABA+ (arrow, insert in I). Olig2-Cre lineage tracing analysis is also performed in the cerebellum of ROSA26;tdTomato (J–P) reporter line at P22. Labeled cells with different morphology in the cerebellum of Olig2-Cre;tdTomato mice are revealed by their tdTomato expression (J). The identity of tdTomato+ cells are examined by co-staining with cell-type specific markers Calbindin (K), Parvalbumin (L), GFAP (M), Olig2 (N), CC1 (O) and Pax2 (P). Note that two tdTomato+ cells are co-labeled with Parvalbumin suggestive of small interneurons (arrows in L). DAPI is used to label nuclei. ML, molecular layer; IGL, internal granule layer; WM, white matter; DCN, deep cerebellar nuclei. Scale bar: 100 μm in (A,J); 20 μm in (B–H,K–P); 25 μm in I.