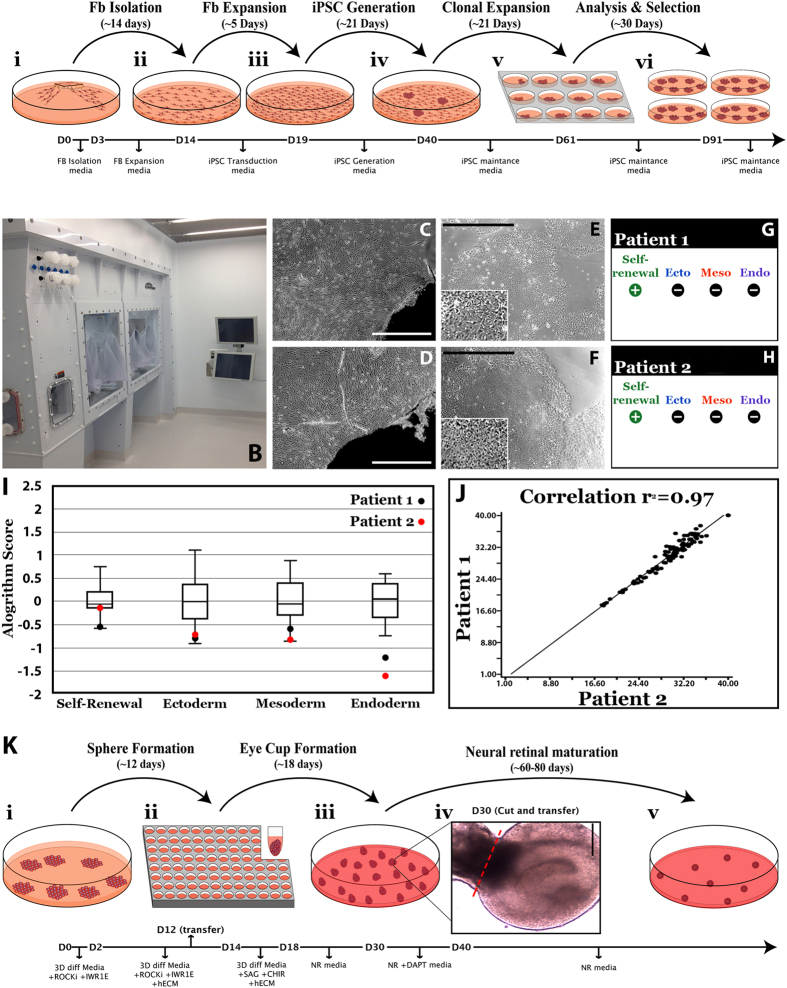
Figure 1. Generation of cGMP, clinical-grade patient-specific iPSCs.
(A) Schematic depicting the timeline and stepwise procedure for cGMP-compliant fibroblast isolation from patient skin biopsies and subsequent generation, clonal expansion and analysis of patient-derived iPSCs. (B) Photograph of one of two cGMP processing suites within the Steven W. Dezii Translational Vision Research Facility equipped with an ISO class 5 BioSpherix Xvivo Closed Incubation System. (C–F) Light micrographs of two independent patient-derived cell lines (DB-005 (C,E) and DB-006 (D,F)). Fibroblasts can be seen migrating from and growing around a fragment of patient skin (C,D). Typical iPSC colonies with large nuclear to cytoplasm ratio generated from each fibroblast line are also shown (E,F and insets in each). (G,H) Data generated by hPSC Scorecard Analysis Software demonstrating each independent patient line primarily expresses genes that participate only in self-renewal, not in formation of endoderm, mesoderm or endoderm. (I) Graph comparing the algorithm scores for expression of genes involved in self-renewal. Compared to the internal reference data set provided by the software, each patient line falls within the average. (J) Graph showing the correlation coefficient comparing all gene expression data for each patient line. The two lines are highly correlated to one another (r2 = 0.97). These data demonstrate how similar each line is to one another, speaking to the consistency of our cGMP protocol for the generation of patient-specific iPSC lines. (K) Schematic showing the timeline and stepwise procedure for cGMP-compliant three-dimensional differentiation of patient iPSCs and the production of iPSC-derived retinal organoids.