Figure 2. Techniques to investigate lncRNA properties and tissue expression.
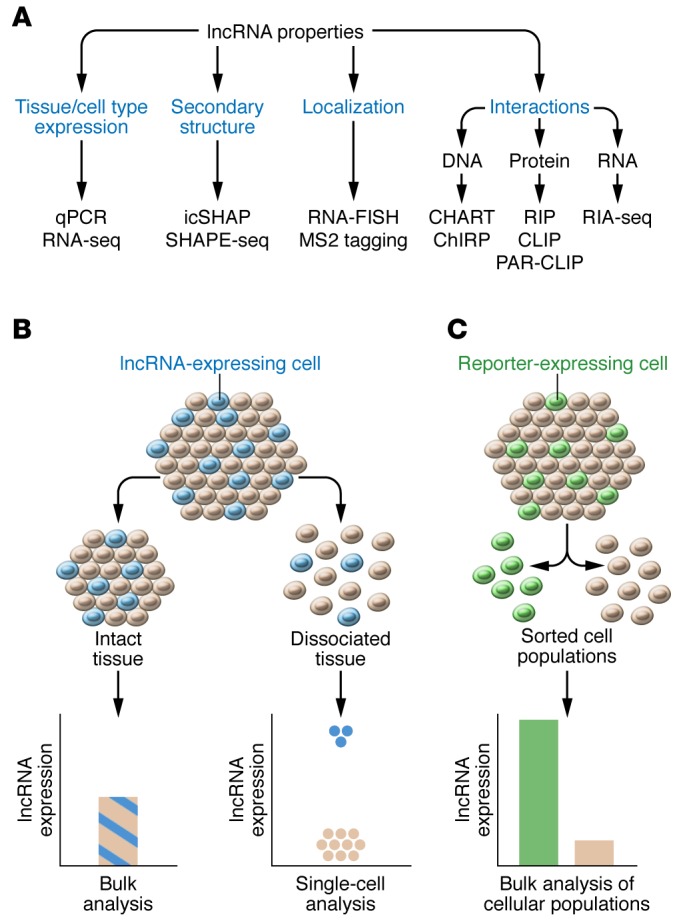
(A) These techniques include quantitative PCR (qPCR) and RNA-seq for transcript expression; RNA fluorescence ISH (FISH) or MS2 tagging (128) for localization; icSHAPE (24) or SHAPE-seq (25) for secondary structures; RIP, CLIP (21, 22), or PAR-CLIP (23) for protein interactions; RIA-seq (20) for RNA interactions; and CHART (18) or ChIRP (19) for DNA interactions. (B) Dissociation of tissue into single cells, followed by single-cell–sequencing analysis, can improve the sensitivity of detection for lncRNA expressed in a minority of cells within a tissue. (C) Fluorescently labeled cells within a tissue can be dissociated, sorted on the basis of fluorescence detection, and assayed for lncRNA expression to improve the sensitivity of detection.
