Fig. 1.
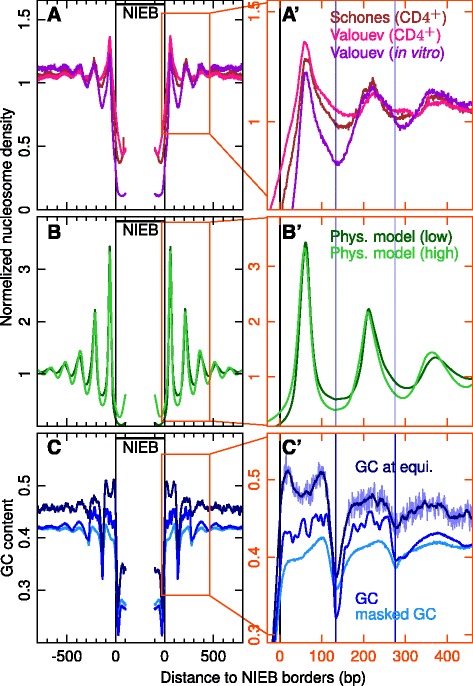
Normalized (with respect to genome average) mean nucleosome density on both sides of the 1,581,256 NIEBs predicted by the sequence-dependent physical model. a “Schones” in vivo (brown), “Valouev” in vivo (pink) and “Valouev” in vitro (purple) data (Methods). b Numerical mean profiles predicted by the physical model at low (dark green) and high (light green) genomic nucleosome coverages (Methods). c Mean GC content (blue), repeat-masked GC content (sky blue) and GC content at equilibrium (navy blue). a’,b’,c’ are zooms on the profiles in (a,b,c) on the right-hand side of the in silico NIEBs ; vertical blue lines correspond to local minima of the GC content (they will be reported in Figs. 4, 5, 6 and 7 for comparison). All profiles were computed at 1 bp resolution, except the ‘GC at equilibrium’ which was smoothed over 10 bp windows (its non-smoothed profile is shown as a background in (c’))
