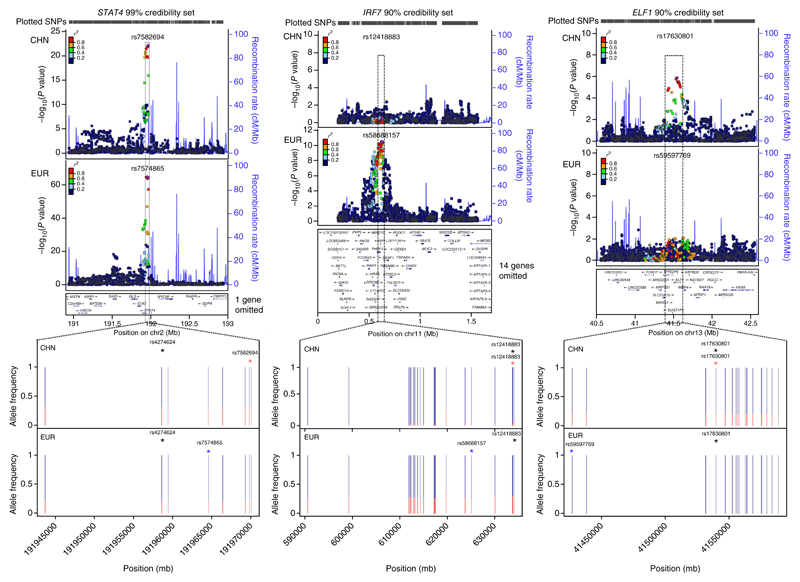
Figure 2.
Fine-mapping examples for STAT4, IRF7 and ELF1. The upper plots are LocusZoom plots showing association significance (−log10(P value)) and local LD (r2; color-coded). Circular points represent SNPs contained within the credibility sets, and square points represent SNPs not contained in the sets. The lower plots display the minor allele frequencies for all the SNPs in the intersection of the European (EUR) and Chinese (CHN) credibility sets. The minor allele frequency is plotted in red. The SNPs with the highest posterior probability within the intersection of the confidence intervals are highlighted by blue (highest posterior probability in the EUR data), red (highest posterior probability in the CHN data) and black (highest posterior probability in the CHN–EUR meta-data) asterisks. The credibility set coverage (99% for STAT4, 90% for IRF7 and ELF1) was chosen as the maximum coverage that included a maximum of 30 SNPs.