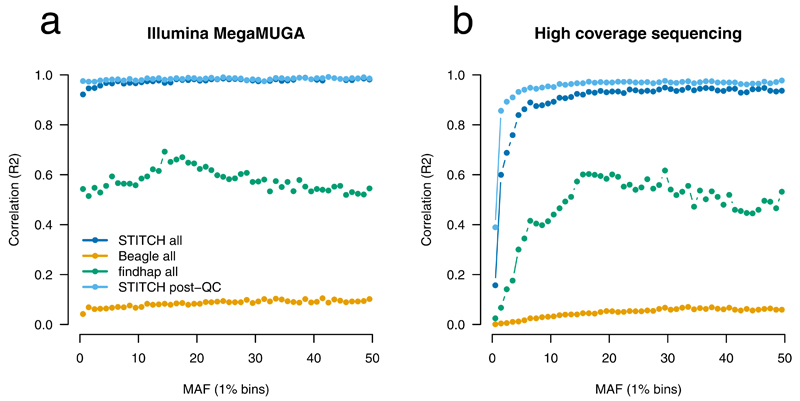
Figure 2. Performance of STITCH on CFW mice compared to external validation.
Validation dataset is the Illumina MegaMUGA array (a) and 10X Illumina sequencing (b). Results are shown for STITCH (K=4, diploid mode), Beagle (default) and findhap (maxlen=10000, minlen=100, steps=3, iters=4) genome-wide for n=2,073 mice featuring 7.07M SNPs before QC and 5.72M after QC. STITCH is run using K=4, diploid method, 40 iterations. Post-QC is SNPs with info>0.4 and HWE p-value > 1×10−6.