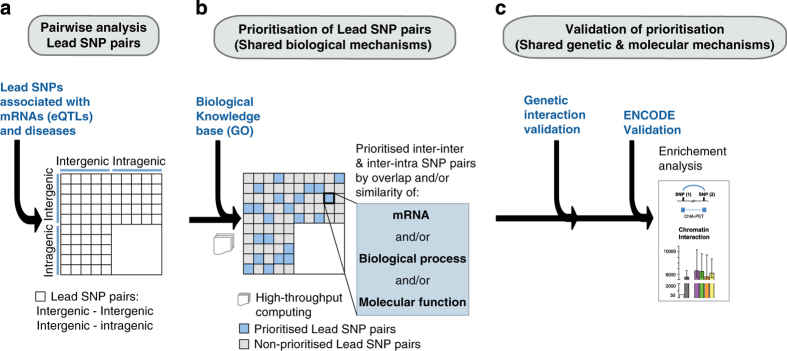
Figure 1.
Schematic of Lead SNP pair prioritisation methods. (a) Lead SNP pairs analysed in this study contain at least one intergenic SNP and are associated with one or more of 467 diseases in the NHGRI GWAS catalogue and with gene expression levels (6,301 mRNAs) derived from a lymphoblastoid cell line eQTL study. Although computed, pairs consisting of two intragenic SNPs were not the main focus of this study (blank in matrix). (b) Lead SNP pairs were prioritised and controlled with empirical scale-free networks to yield significant Lead SNP pairs sharing at least one of the three imputed biological mechanisms (blue highlighted squares). Biological knowledge bases refer to eQTL associations and gene ontology annotations of molecular functions and biological process. (c) Prioritised inter–inter and inter–intra Lead SNP pairs were further validated for genetic interaction using three independent association studies (GWAS and PheWAS), and for shared TFs and interacting regulatory elements using ENCODE-derived data sets.