Figure 2.
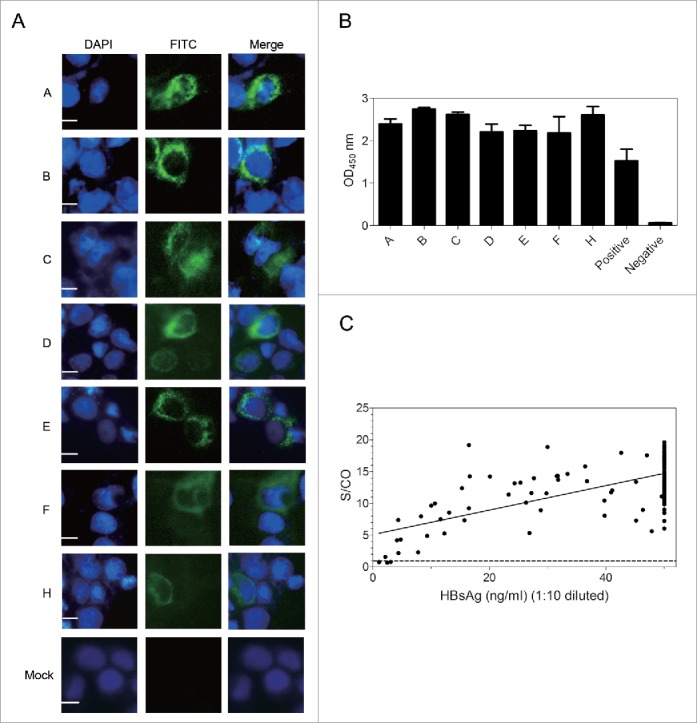
G12 can recognize HBsAg from different HBV genotypes and vast majority of clinical samples. ((A)& B) Huh7 cells were transfected with 1.3 copies of the HBV genome belonging to genotype A-F and H, followed by IF staining of intracellular envelope proteins by G12 (A) and ELISA detection of HBsAg released to culture supernatant using G12 coated plate (B). The three images for panel A are, from left to right, nuclear staining by DAPI (blue), HBsAg staining by G12 followed by FITC-conjugated anti-human antibody (green), and merged images. Each bar represents 10μm. Cells transfected with pUC18 DNA served as a negative control (Mock). (C) G12-based ELISA to detect HBsAg from 198 serum samples. The horizontal axis shows amounts of HBsAg (ng/ml) based on Light Initiated Chemiluminescence Analyzing System (Bo Yang), while vertical axis represents values (S/CO) from ELISA with G12-coated plates. For G12-based ELISA the cutoff value was 2.1 times the average of negative controls, and samples with S/CO >1 were scored positive (dotted line). S/CO: signal over cutoff.
