Figure 6.
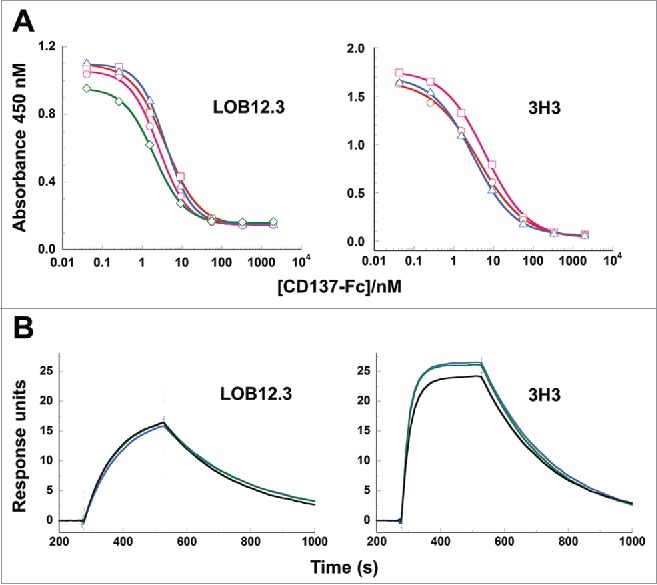
Binding of reverse-engineered LOB12.3 and 3H3 Fab variants to murine CD137. A) Competition phage ELISA measuring relative CD137 binding affinities to phage displayed LOB12.3 or 3H3 Fab variants. LOB12.3 Fab variants contained LOB12.3-Lv3/Hv3 light/heavy V-region pairs, where the sequence at VL33/VH100c was Leu/Leu (green), Leu/Ile (blue), Ile/Leu (magenta) or Ile/Ile (red). IC50 values for displacement of 50% of bound Fab-phage was between 2.0 – 3.9 nM for all 4 variants. 3H3 Fab variants contained 3H3-Lv2/Hv3 V-region pairs, where the sequence at VH29 was Ile (blue), or contained a VH-E61D substitution and Ile (red) or Leu (magenta) at VH29. IC50 values for displacement of 50% of bound Fab-phage was between 3.1 – 6.2 nM for all 3 variants. B) Sensorgram traces showing near identical binding kinetics for 11.1 nM injections of Fab samples over an immobilized CD137 surface. Reverse-engineered Fab variants (blue = Fab1; green = Fab2) are compared to the corresponding reference Fabs generated by partial proteolysis of the commercially-sourced parental IgG (black = Fabp). Binding kinetics calculated from the full set of surface plasmon resonance data are shown in Table 3.
