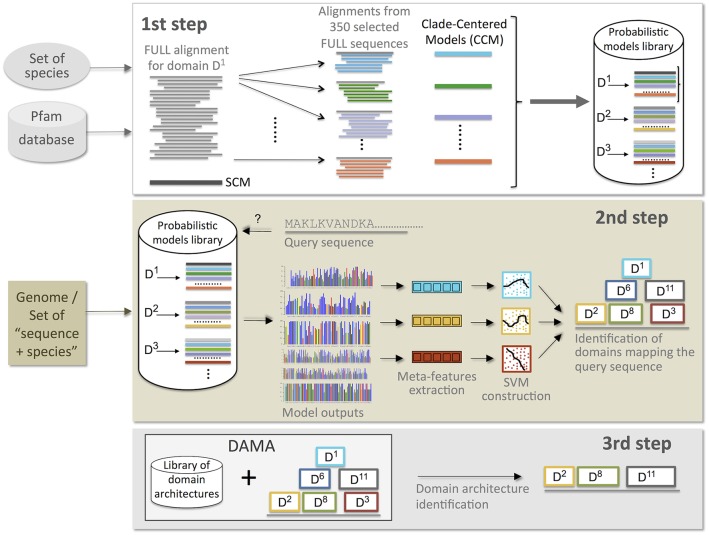
Fig 1. Schema representing the three main steps of CLADE method.
The 1st step (top) concerns the construction of domain profiles from the Pfam database. A specific set of species can be furnished to CLADE (optional) to guide the selection of homologous sequences (and species) for the construction of clade-centered models (CCM), otherwise set to be a random selection. The output is a library of probabilistic models: for each Pfam domain, it contains a SCM, provided by Pfam, and a large number of CCMs, associated to the FULL set of Pfam sequences for the domain. All probabilistic models are used to identify potential domains occurring in query sequences. The schema illustrates the model construction for domain D1; it is applied to all Pfam domains. The 2nd step (middle) matches all models generated in step 1 against query sequences belonging to a given genome or to a set of sequences given as input, and identifies a set of potential domains occurring in the sequences. Then, it filters potential domains by using support vector machines. For each domain, it constructs a SVM that combines multiple features extracted from the SCM and CCM models associated to the domain. The schema illustrates domain identification for a given query sequence; it is applied to all input sequences. The 3rd step (bottom) takes the position of potential domains in a query sequence (from step 2) and runs DAMA, a tool designed to predict domain architectures from known ones.