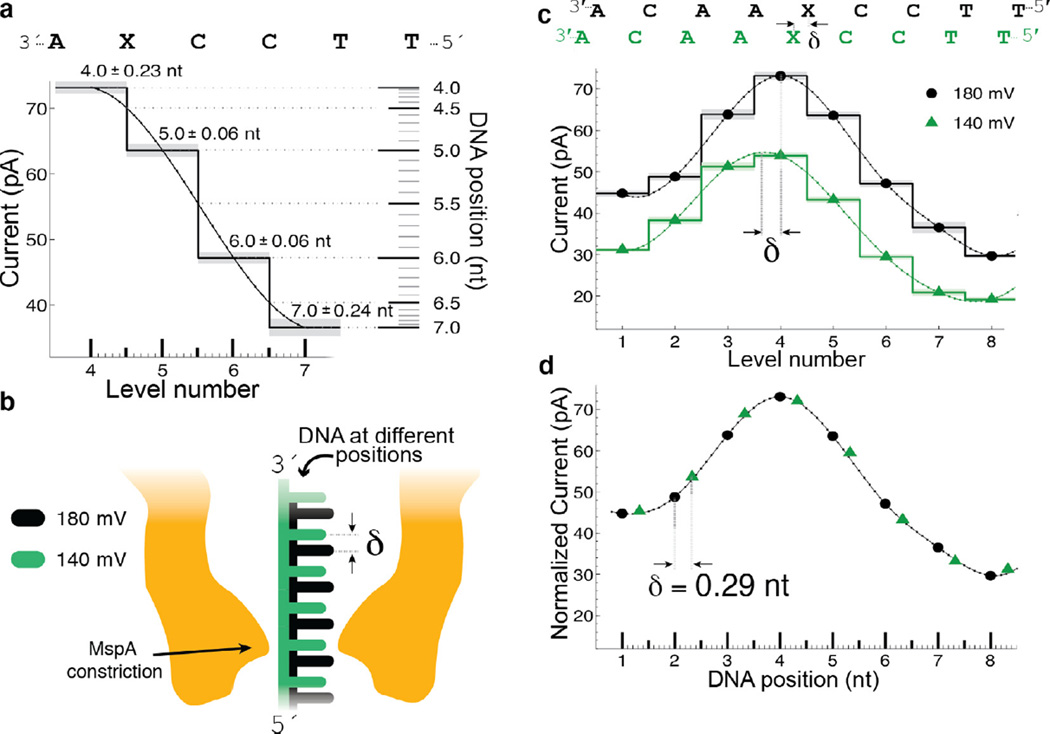
Fig. 6.
Transduction of current to distance. (a) Regions of high current contrast can be used to measure DNA position precisely. Small uncertainties in measured current translate to small positional uncertainty. (b) Schematic depiction of DNA position within the pore at two different voltages; differences in the applied electric force result in different DNA extensions. (c) Current levels observed for phi29 DNAP controlled motion of DNA through MspA at 180 mV and 140 mV of applied potential. A cubic spline interpolant has been applied to each set of current steps. Note that, apart from a scaling factor, the shape of the spline is identical but the position of the spline has shifted a distance δ. (d) After a linear scale and offset is applied to the two splines, a horizontal displacement δ = 0.29 nt brings the two splines in line with one another. This experiment has two important results: (1) The current levels observed during single-nucleotide stepping of DNA through MspA lie along an underlying smooth curve that is well-approximated by a spline. (2) This spline provides a direct mapping from current to DNA position and we can use it to measure sub-nucleotide movement of DNA. Figure modified from [37].