Figure 3. Generating combinatorial barcode diversity in transgenic zebrafish.
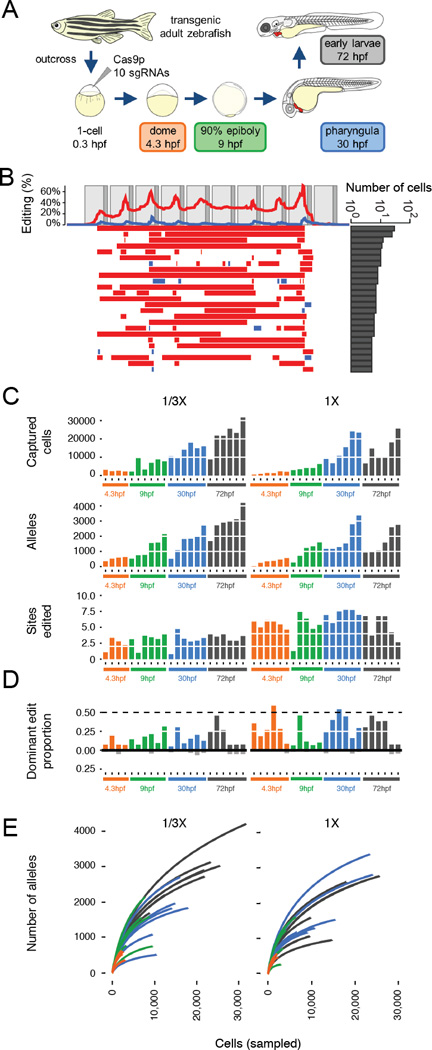
(A) One-cell zebrafish embryos were injected with complexed Cas9 ribonucleoproteins (RNPs) containing sgRNAs that matched each of the 10 targets in the array (v6 or v7). Embryos were collected at time points indicated. UMI-tagged barcodes were amplified and sequenced from genomic DNA. (B) Patterns of editing in alleles recovered from a 30 hpf v6 embryo, with layout as described in the Fig. 1B legend. (C) Bar plots show the number of cells sampled (top), unique alleles observed (middle) and proportion of sites edited (bottom) for 45 v7 embryos collected at four developmental time-points and two levels of Cas9 RNP (1/3x, 1x). Colors correspond to stages shown in panel (A). Although more alleles are observed with sampling of larger numbers of cells at later time points, the proportion of target sites edited remains relatively constant. (D) Bar plots show the proportion of edited barcodes containing the most common editing event in a given embryo. Six of 45 embryos had the most common edit in approximately 50% of cells (dashed line), consistent with this edit having occurred at the two-cell stage (see fig. S8A for example). Colors correspond to stages shown in panel (A). These same edits are rarer or absent in other embryos (black bars below). (E) For each of the 45 v7 embryos, all barcodes observed were sampled without replacement. The cumulative number of unique alleles observed as a function of the number of cells sampled is shown (average of the 500 iterations shown per embryo; two levels of Cas9 RNP: 1/3x on left, 1x on right). The number of unique alleles observed, even in later developmental stages where we are sampling much larger numbers of cells, appears to saturate, and there is no consistent pattern supporting substantially greater diversity in later time-points, consistent with the bottom row of panel (C) in supporting the conclusion that the majority of editing occurs before dome stage.
