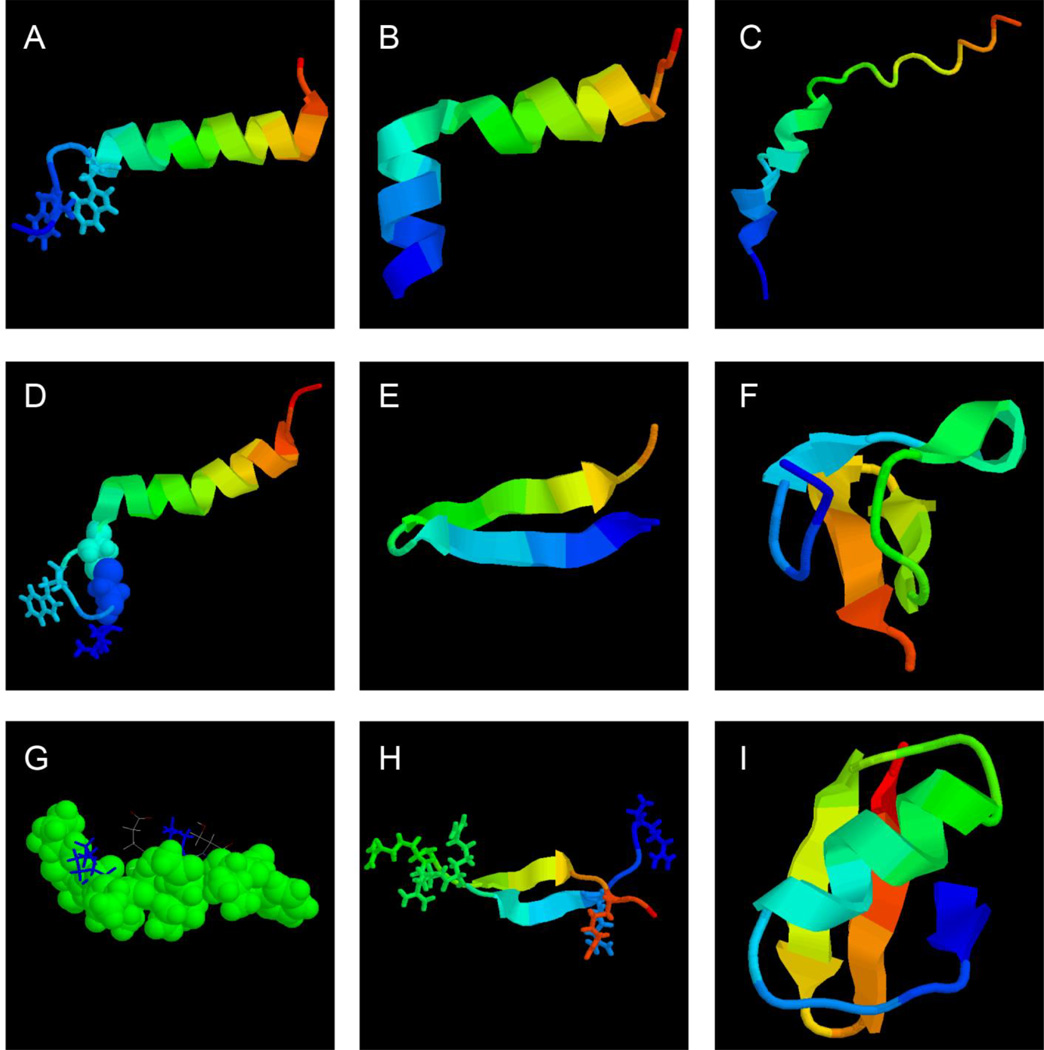
Figure 1.
Structural biology of spider (A–F) and scorpion (G–I) antimicrobial peptides. Shown are the NMR structure of spider (A) latarcin 1 (PDB ID: 2PCO), (B) latarcin 2a (PDB ID: 2G9P), (C) cupiennin 1a (PDB ID: 2K38), (D) oxyopinin 4a (PDB ID: 2L3I), (E) gomesin (PDB ID: 1KFP), (F) psalmopeotoxin I (PDB ID: 1×5V), scorpion (G) IsCT (PDB ID: 1T51), (H) androctonin (PDB ID: 1CZ6), and (I) charybdotoxin (PDB ID: 2CRD), respectively. In these structural views, the peptide chain is colored from the N (blue) to the C-terminus (red). To emphasize the broad hydrophobic surface of IsCT in panel G, as well as the hydrophobic nature of sulfurs involved in the disulfide bond in panel D, they are displayed in space-filling models (G). The figure was generated using the software RasMol (https://www.umass.edu/microbio/rasmol/). See the text for further details.