Fig. 5.
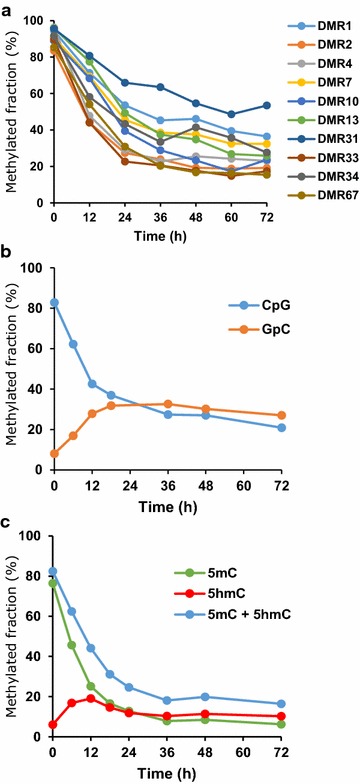
Time-course analysis. a Demethylation of 10 DMRs. Average CpG methylated fractions in Hm06 donor monocytes (0 h) and cells collected at different time points during differentiation into macrophages. Related to Additional file 9: Figure S5. b Time course of chromatin accessibility (GpC methylation in NOMe experiments) and DNA CpG methylation in DMR33. Average CpG and GpC methylated fractions in Hm10 donor monocytes (0 h) and cells collected at different time points during differentiation into macrophages. The transition from lower to higher GpC methylated fraction is indicative of an increase in chromatin accessibility. GCG motifs were excluded due to ambiguity between CpG- endogenous- and GpC-enzymatic-methylation. Related to Additional file 10: Figure S6. c 5-hydroxymethyl-cytosine (5hmC) level increases during differentiation. Average 5hmC and 5mC (5-methylcytosine) fractions in monocytes (0 h) and cells collected at different time points (DMR33 from donor Hm10). Related to Additional file 14: Figure S7
