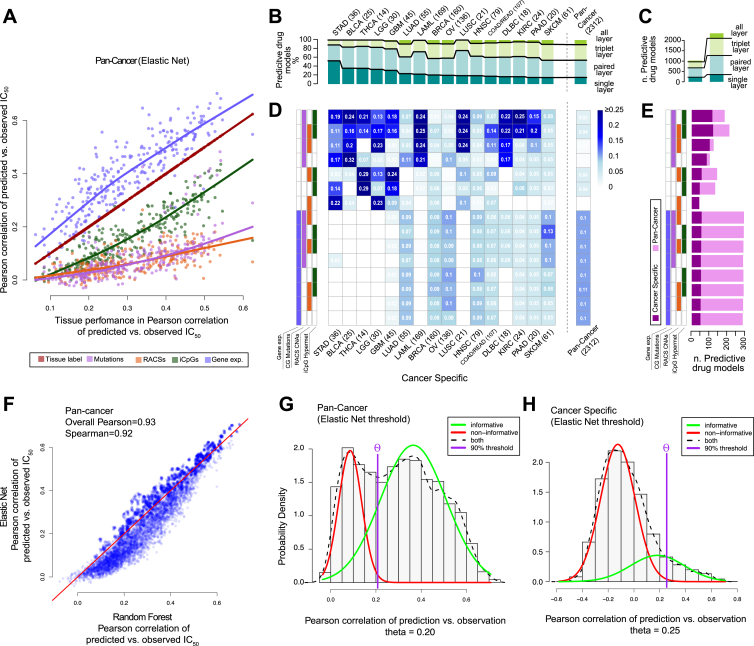
Figure S6.
Predictive Ability Assessment of Individual Molecular Feature Layers and Layer Combinations, Related to Figure 6
(A) Predictive performance (Pearson correlation of predicted versus observed IC50 values) of tissue label versus other feature layers in pan-cancer analysis with Elastic Net.
(B) Percentages of all the predictive models ( and ) across different cancer types and molecular data type. Absolute counts of best performing models are indicated above the bars.
(C) Absolute counts of pan-cancer and cancer-specific models separated by number of feature layers.
(D) Heatmap split by cancer types and possible feature combination, showing the percentage of all predictive models.
(E) Count of all predictive models by data type combination separated in pan-cancer and cancer-specific analysis.
(F) Comparison of Random Forests versus Elastic Net performances in the pan-cancer analysis.
(G) Deriving pan-cancer threshold of predictive models by fitting a mixed Gaussian distribution across all build models, while assuming that one distribution is informative and the other one is not. A model is considered predictive if the ratio of informative to non-informative is at least 9, resulting in a minimal Pearson correlation of ∼0.2 pan-cancer models achieving high performances due to tissue bias.
(H) Deriving cancer-specific threshold in same manner as for pan-cancer, resulting in minimal Pearson correlation of ∼0.25. Negative correlations result from overfitting and too small sample sizes.