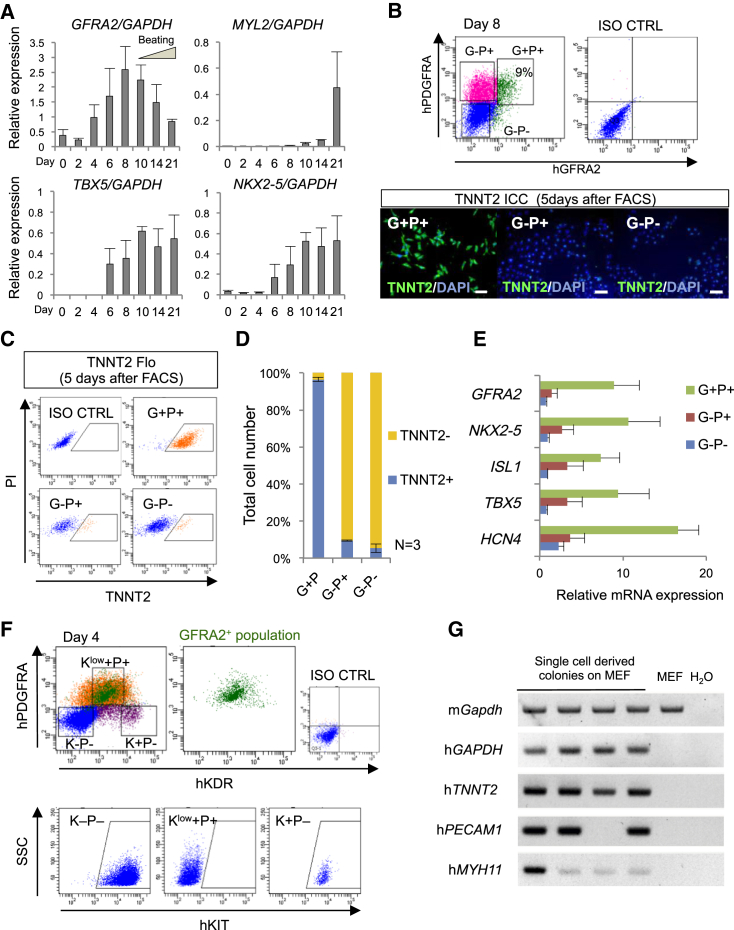
Figure 4.
Human GFRA2 Marks CPs from Human ESC Cultures
(A) qPCR analyses for human GFRA2, NKX2-5, TBX5, and MYL2. Note the peak of human GFRA2 expression is just before human embryoid bodies start to beat. Bar graph represents biological triplicates with technical duplicates as mean ± SEM.
(B) FACS isolation of human hGFRA2+/hPDGFRA+ (G+P+) cells. ICC analyses for TNNT2 demonstrate that isolated G+P+ population is highly cardiomyogenic. Scale bar, 100 μm.
(C and D) Quantitative analyses by Flo for TNNT2 show most of G+P+ cells (96.5% ± 1.1%) differentiated into cardiomyocytes. Bar graph represents mean ± SEM. n = 3.
(E) qPCR analyses for GFRA2, NKX2-5, ISL1, TBX5, and HCN4 in FACS-isolated populations. All cardiac marker gene expressions were significantly higher in the G+P+ population when compared to the G−P− population (∗p < 0.05, Student’s t test). Data are representative of biological triplicates with technical duplicates as mean ± SEM.
(F) Flow cytometry of human ESCs at differentiation day 4. hGFRA2+ cells (shown as green dots) were mostly included by hPDGFRA+/hKDRlow+/hKITneg population, which is reported as multipotent CPs (Yang et al., 2008). 15%–20% of hPDGFRA+/hKDRlow+/hKITneg cells were hGFRA2+.
(G) RT-PCR analyses of single-cell-derived colonies for each lineage marker gene. A single hGFRA2+/hKDRlow+/hPDGFRA+ cell at day 4 was clonally sorted and cultured on mouse embryonic fibroblasts (MEFs) for 2 weeks. RT-PCR of hTNNT2, hPECAM1, and hMYH11 represent the existence of cardiomyocytes, endothelial cells, and smooth muscle cells among the cells derived from a single cell, respectively. Note the existence of multiple lineages, which indicates the multipotency.