Figure 1.
New Genetic Codes
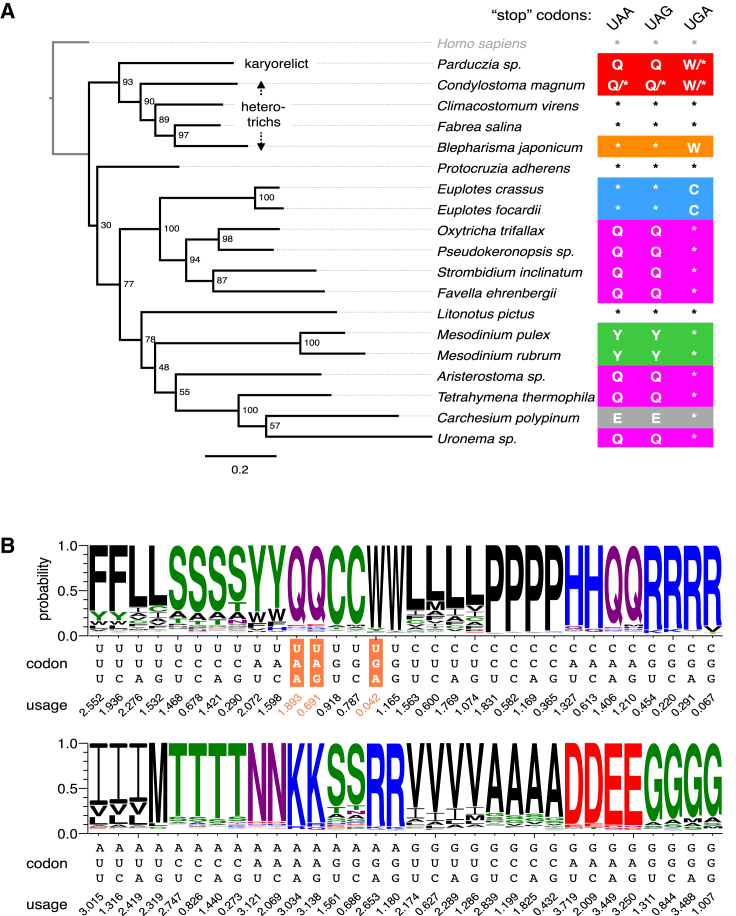
(A) Stop codon reassignments (Q, glutamine; W, tryptophan; C, cysteine; Y, tyrosine; ∗, stop) are mapped onto an eRF1 maximum likelihood phylogeny. Homo sapiens (standard genetic code) is an outgroup. Bootstrap support for every node is shown. Scale bar indicates amino acid substitutions per site. UGA codons were previously found in the coding sequences of Blepharisma americanum and were predicted to encode tryptophan (Eliseev et al., 2011, Lozupone et al., 2001). Experimental assays in Blepharisma japonicum suggest its eRF1 recognizes all three standard stop codons (Eliseev et al., 2011). It should be noted that ciliates from the family Mesodiniidae have both a unique genetic code (UAG/UAA = UAR = tyrosine; UGA = stop) and extremely divergent rRNAs (Johnson et al., 2004).
(B) Predicted C. magnum genetic code. Stop codons are highlighted in orange. Predicted amino acids are those with maximal heights. Codon usage inferred from translated BLAST matches is shown below the codons. UAA and UAG codons were previously predicted to encode glutamine (Lozupone et al., 2001, Tourancheau et al., 1995).