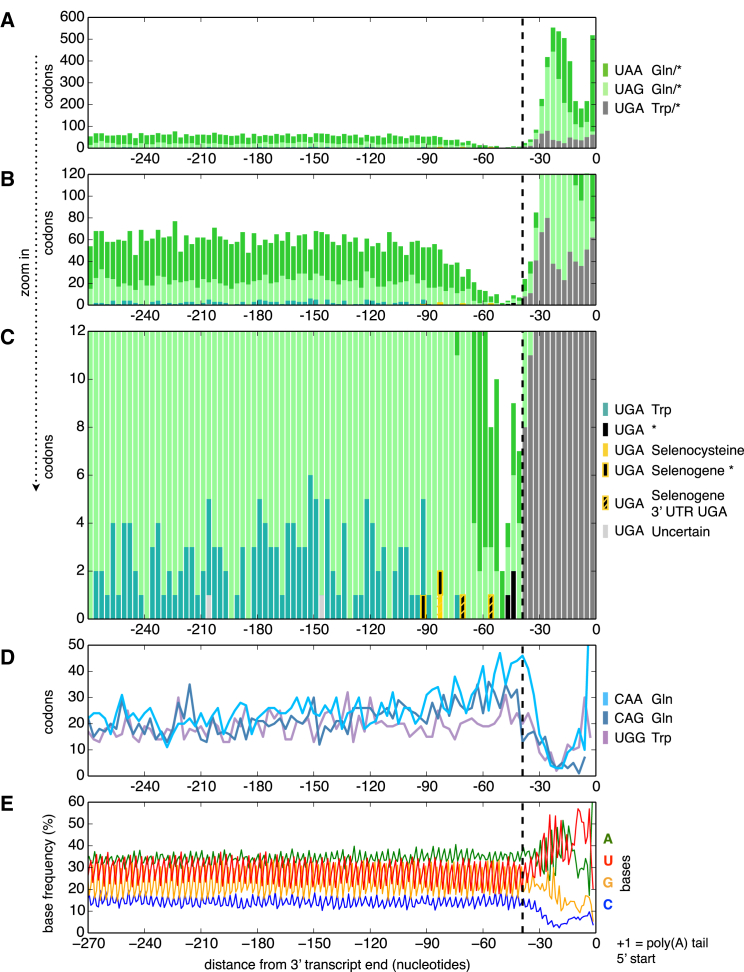
Figure 6.
Terminal “Stop” Codon Decline Close to C. magnum Stops
Stacked bar graphs of “stop” codon counts are for the transcript regions upstream of poly(A) tails (position 0). Transcript ends include 0, 1, or 2 nucleotides of the poly(A) tail to complete the final “codon.” 3′ UTRs occur in the region to the right of the right-most dashed vertical line. Codons counted are those in the 1672 poly(A)-tailed single gene, single isoform Trinity assembled transcripts.
(A–C) The top three subgraphs are drawn in decreasing order of ordinate limits. Vertical line at −39 nt indicates approximately where most downstream “stops” are either stop codons or “codons” in 3′ UTRs. Codons whose sense/stop states have not been determined are indicated by “amino acid/∗.” Transcripts with UGA codons upstream of −39 nt were visually classified based on BLASTX searches. Upstream of −39 nt, UGA codons predominantly code for tryptophan; downstream of −39 nt, UGA codons are predominantly stops or codons in 3′ UTRs downstream of primary stops (both indicated by gray bars). In the genetic codes of C. magnum and Parduczia sp. UGA is a codon triality (codon duality is reviewed in Atkins and Baranov, 2007), because in addition to being interpreted as a tryptophan codon and a stop codon, it also serves as a selenocysteine codon in the context of SECIS elements. Pale gray bars correspond to a transcript with an uncertain C-terminal, as judged by BLAST.
(D) Standard glutamine and tryptophan sense codon counts.
(E) Base frequencies are stable in the region of “stop” codon decline (∼−90 to −42 bases upstream of poly-As).
See also Figures S5 and S6.