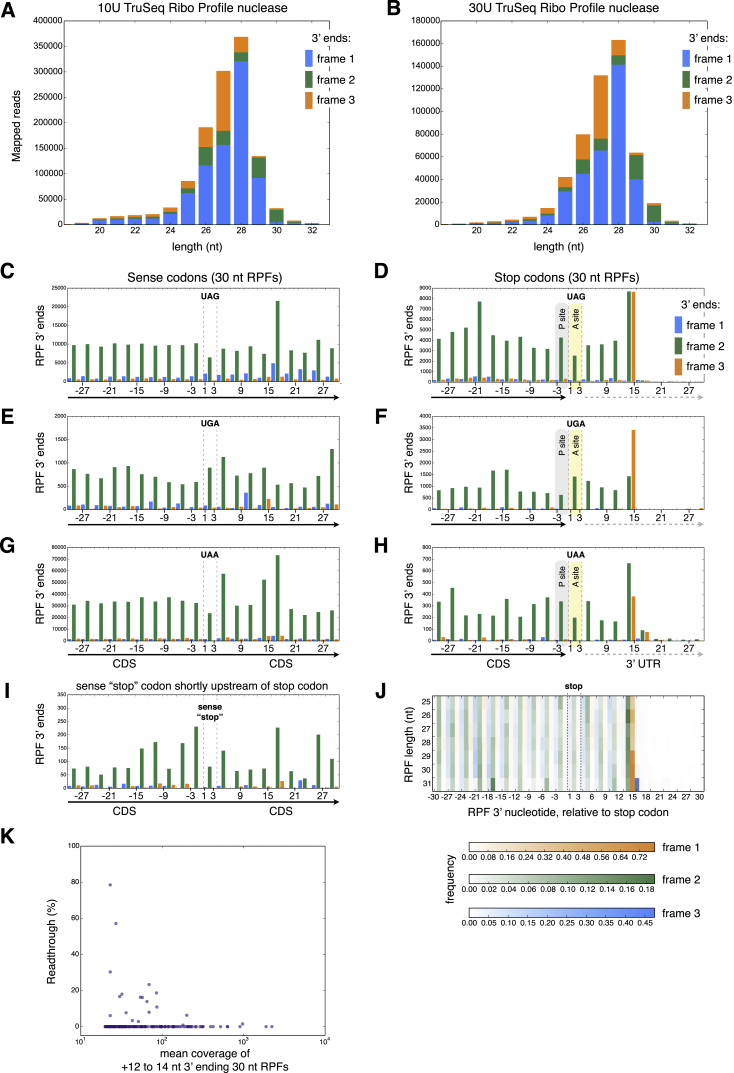
Figure S3.
Properties of Ribo-seq Data at Sense and Stop Codons, Related to Figure 3
(A and B) Distributions for 10U and 30U of TruSeq Ribo Profile nuclease used to produce RPFs. The peak RPF length is at 28 nt and most RPF 5′ starts and 3′ ends are in frame 1 as for Saccharomyces cerevisiae RPFs (Ingolia et al., 2009).
(C–H) Distribution of 30 nt RPFs for individual sense and stop UAG, UGA and UAA codons (positions 1 to 3) in Trinity assembled transcripts.
(I) 30 nt RPF coverage of UAA, UGA and UAA codons located 24-66 nucleotides upstream of their stops.
(J) RPF 3′ end distribution around stop codons for 25-31 nt RPFs; frequencies of RPF ends are calculated for each RPF length.
(K) Stop codon readthrough. See the Supplemental Experimental Procedures for the manner in which readthrough was measured.