Figure 2.
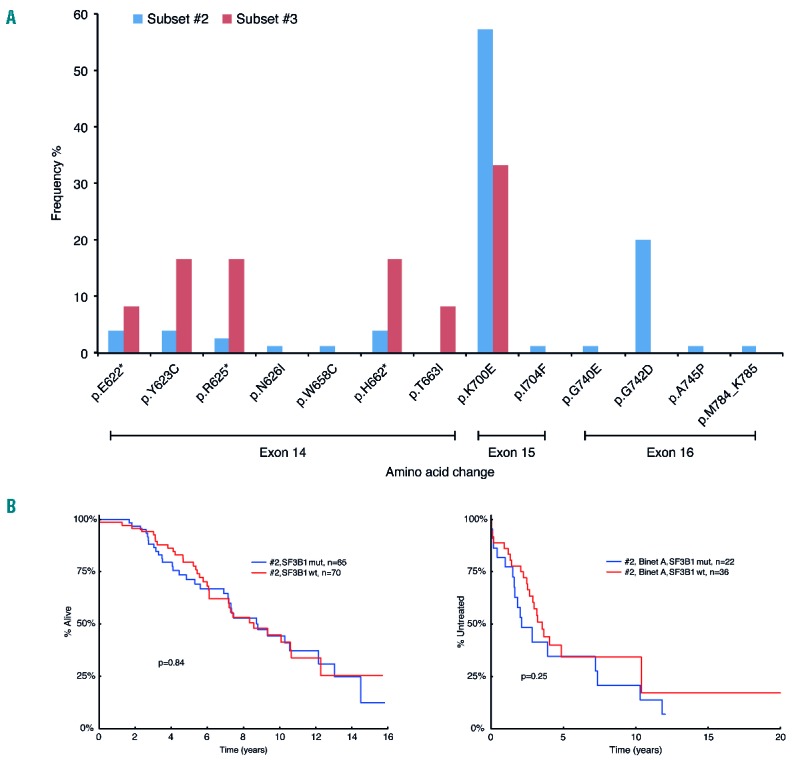
SF3B1 mutations in subsets #2 and #3. (A) Distribution of SF3B1 mutations in subsets #2 and #3. Overall, 45% (72/161) of subset #2 and 46% (12/26) of subset #3 cases were found to carry mutations within SF3B1. While the majority of subset #2 cases (69/72; 96%) carried a single SF3B1 mutation, 3 cases had 2 mutations within SF3B1 (2/3 cases carried the p.K700E change, with one case carrying both p.K700E and p.G742D). Although the most frequent amino acid change in both subsets involved a lysine to glutamic acid substitution at codon 700 (exon 15), representing 57% (43/75) and 33% (4/12) of all SF3B1 mutations in subsets #2 and #3, respectively, the overall distribution of mutations varied. To elaborate, in subset #3, all remaining SF3B1 mutations (excluding p.K700E) occur within exon 14. This is in contrast to the SF3B1 mutational profile in subset #2 where only 17% of mutations are found within exon 14 and where two particular substitutions account for 96% and 88% of the alterations observed in exon 15 and exon 16 (p.K700E and p.G742D, respectively). *indicates that more than one amino acid change occurred at this codon (detailed in Online Supplementary Table S8). (B) Prognostic implications of SF3B1 mutations within subset #2 on overall survival (OS) and time to first treatment (TTFT). (Binet A cases).
