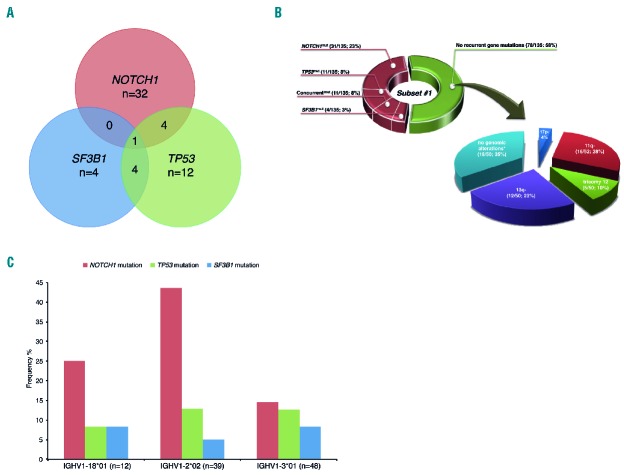
Figure 3.
Main biological associations within subset #1. (A) Concurrent mutations within subset #1. Only cases for which the mutational status of all 3 genes (NOTCH1, SF3B1 and TP53) was available were included in the figure (n=135). Fifty-seven cases had a mutation in at least one of the gene hotspots whereas 78 cases were wild-type for these genes. (B) Spectrum of mutations and genomic aberrations within subset #1. Despite a high frequency of NOTCH1 mutations, a large proportion (58%) of subset #1 cases carried no mutations within the 5 genes analyzed. Specifically, considering the subset #1 cases lacking any recurrent gene mutations, 35% also lacked any recurrent genetic aberrations. Collectively, this resulted in the absence of any recurrent gene mutation or cytogenetic aberration in approximately 20% of subset #1 cases, thereby implying that additional mechanisms must account for the clinically aggressive nature of this subset. Only cases for which the mutational status of all 3 genes (NOTCH1, SF3B1 and TP53) was available were included in the figure (n=135). *indicates that none of the known recurrent genomic aberrations were present; NOTCH1mut: mutation in NOTCH1 only; TP53mut: mutation in TP53 only; SF3B1mut: mutation in SF3B1 only. Concurrentmut refers to the presence of mutations in more than one of the genes analyzed. Absolute numbers and percentages are provided in brackets. For del(17p), 2/53 correspond to the 4% indicated in the figure. (C) The frequency of NOTCH1 mutations in subset #1 cases varies depending on specific IGHV gene usage. Only the top 3 utilized IGHV genes within subset #1 patients in our cohort were included in the graph, collectively accounting for 73% (99/136) subset #1 cases. Mutations within NOTCH1 were found to be particularly frequent in subset #1 cases expressing IGHV1-2*02 (17/39; 44%).