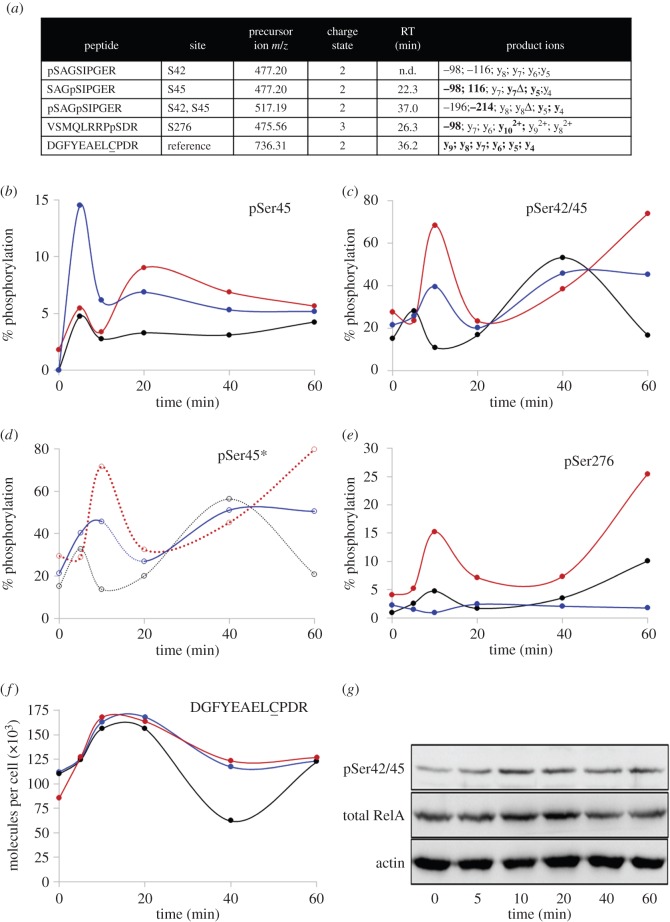
Figure 4.
Selected-reaction monitoring (SRM) of RelA phosphorylation in response to treatment of SK-N-AS cells with the cytokine TNFα. (a) List of targeted (phospho)peptides included in the SRM analysis. Sequences of peptides, including phosphorylation sites (p), are listed along with their respective precursor ion m/z values, precursor ion charge state, retention time (RT) and product ions. C represents carbamidomethylated cysteine; −98 indicates loss of H3PO4 from the precursor ion; −116 indicates loss of (H3PO4 + H2O) from precursor ion; n.d. not detected; consistently observed product ions are highlighted in bold. (Phospho)peptides containing modified residues pSer45 (b), pSer42/45 (c), pSer45* (d), pSer276 (e) and the RelA control peptide DGFYEAELCPDR, where C denotes carbamidomethylation of Cys (f) were quantified by scheduled LC–SRM analysis following stimulation of SK-N-AS cells with TNFα; stimulation and analysis was repeated in triplicate for three different biological replicates represented in black, blue and red. Isotope-labelled internal reference (phospho)peptides were included, permitting absolute peptide quantification and phosphorylation site stoichiometry determination. pSer45* is the calculated stoichiometry of the Ser45 phosphorylation site obtained by summating percentage phosphorylation of the pSer45 and pSer42/45 phosphopeptides. Immunoblot analysis of pSer42/45 (g) is shown as a function of time following TNFα stimulation with reference to total RelA levels and actin control.