Figure 1. RBR E3 ligases do not require closed states of E2~Ub for ubiquitin transfer.
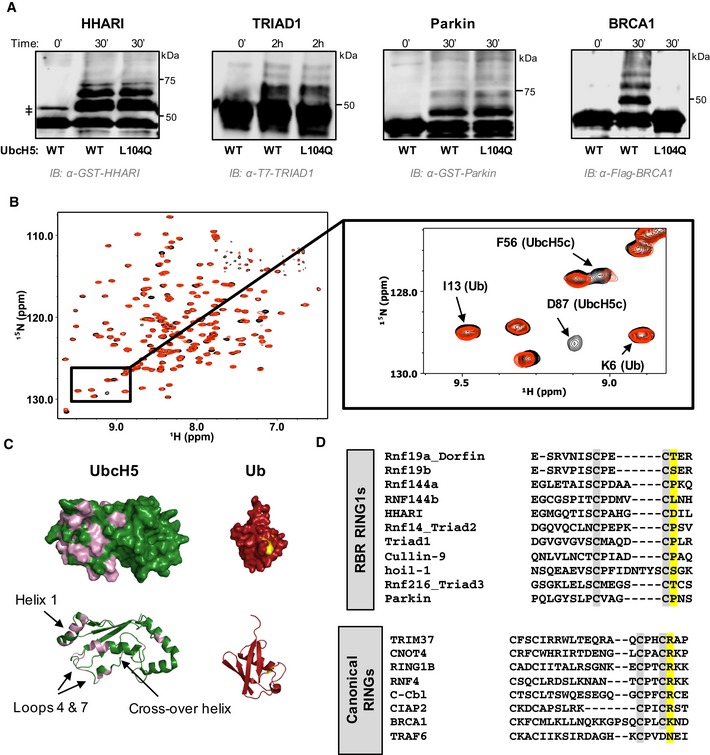
- Auto‐ubiquitination assays in which the E3 acts both as an E3 and as proxy substrate were performed with GST‐HHARIRBR, T7‐Triad1ΔAri, GST‐ParkinRBR , and Flag‐BRCA1/BARD1 and either UbcH5WT or UbcH5L104Q as the E2. Products were visualized by Western blotting against the indicated tags on E3s. Times given are post‐ATP addition.
- Overlay of (1H,15N)‐HSQC‐TROSY spectra of 15N‐UbcH5‐O‐15N‐Ub in the absence (black) and presence (red) of 0.5 mol equiv. HHARI RING1. A subset of UbcH5 peaks, but not Ub peaks, shift and broaden upon HHARI RING1 binding.
- Chemical shift perturbations (CSPs) from (B) are mapped onto the structures of UbcH5c (PDB 2fuh) and Ub (PDB 1ubq). Residues that exhibit loss of intensity > 1 stdv upon RING1 binding (intensity < 0.47) are colored on each structure: UbcH5 residues 5, 6, 7, 16, 20, 22, 56, 62, 74, 87, 90, 91, 96, 97, 99–103, 137, 138 (pink) and Ub residues 48, 50 (yellow). The lack of CSPs on the UbcH5 crossover helix and on the surface of Ub signifies that HHARI RING1 does not induce closed UbcH5˜Ub.
- CLUSTAL OMEGA alignments of the C‐terminal sequences of RBR RING1 domains (top) and canonical RING domains (bottom). Each alignment includes the 7th and 8th Zn2+ coordinating positions (gray). The allosteric linchpin position critical for E2˜Ub activation by RING‐type E3s is highlighted in yellow. RBR RING1 domains lack the allosteric linchpin.
