Fig. 4.
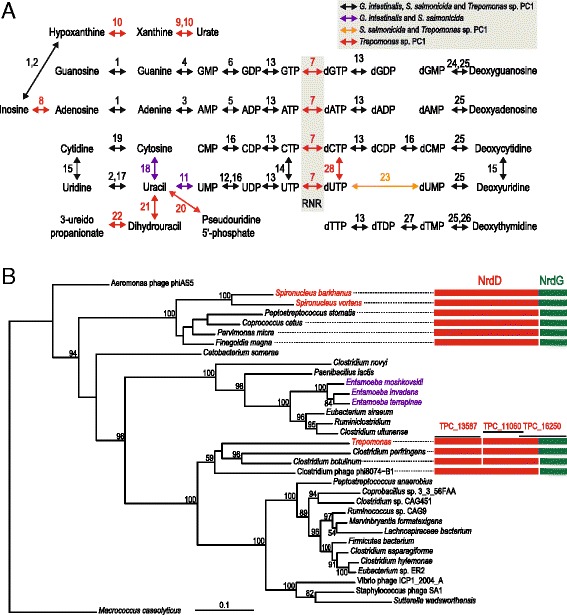
Nucleotide metabolism in diplomonads. a Proposed pathways for synthesis and scavenging of purine ribonucleosides, pyrimidine ribonucleosides and deoxynucleosides. The presence of enzymes across G. intestinalis, S. salmonicida and Trepomonas sp. PC1 is indicated by colors. Black: shared by all, purple: G. intestinalis + S. salmonicida, orange: S. salmonicida + Trepomonas sp. PC1, red: Trepomonas sp. PC1 only. Key to enzymes: 1. purine nucleoside phosphorylase, 2. inosine-uridine nucleoside N-ribohydrolase, 3. adenine phosphoribosyltransferase, 4. guanine phosphoribosyltransferase, 5. adenylate kinase, 6. guanylate kinase, 7. anaerobic ribonucleotide reductase (RNR), 8. adenosine deaminase, 9. xanthine dehydrogenase, 10. xanthine oxidase, 11. uracil phosphoribosyltransferase, 12. UMP kinase, 13. nucleoside diphosphate kinase, 14. CTP synthase, 15. cytidine deaminase, 16. UMP-CMP kinase, 17. uridine/thymine phosphorylase, 18. cytosine deaminase, 19. cytidine hydrolase, 20. pseudouridine-5’-phosphate glycosidase, 21. dihydrouracil dehydrogenase, 22. dihydropyrimidinase, 23. deoxyuridine-5’-triphosphate nucleotidohydrolase, 24. deoxyguanosine kinase, 25. deoxynucleosidase kinase, 26. thymidine kinase, 27. thymidylate kinase, and 28. deoxycytidine triphosphate deaminase. b Protein maximum likelihood phylogeny of class III anaerobic RNR of the NrdD class. The arrangement of NrdD and its activating protein NrdG in Spironucleus, Trepomonas and their closest relatives are indicated by boxes. Three Trepomonas transcripts make up a putative RNR. TPC1_13587 covers the N-terminal part of NrdD. TPC1_11060 and TPC1_16250 overlaps each other with 13 amino acids in the C-terminal part of NrdD. TPC1_16250 shows that NrdD is fused with NrdG in Trepomonas. The tree is inferred for NrdD, which is found in a single peptide in all species except Trepomonas and Clostridium botulinum and Clostridium perfringens. Eukaryotes are labeled according to their taxonomic/phylogenetic classification [13]: Amoebozoa (purple) and Excavata (red). Only bootstrap support values > 50 % are shown
