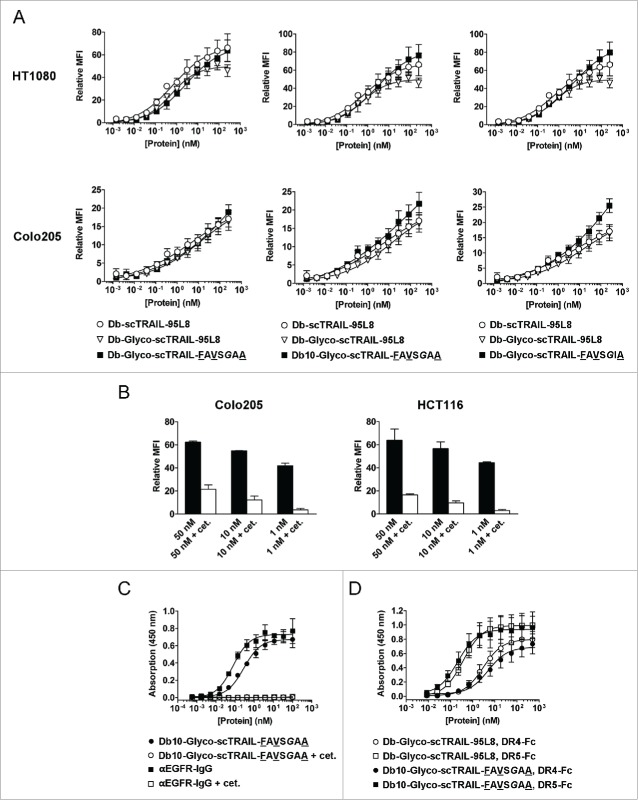
Figure 4.
Binding of EGFR-targeted Db-scTRAIL proteins. (A) The binding of Db-Glyco-scTRAIL-FAVSGAA (left), Db10-Glyco-scTRAIL-FAVSGAA (middle) and Db-Glyco-scTRAIL-FAVSGIA (right, squares) to EGFR+ HT1080 fibrosarcoma (upper) and Colo205 colon carcinoma cells (lower) was measured by flow cytometry in reference to Db-scTRAIL-95L8 (circles) and Db-Glyco-scTRAIL-95L8 (triangles). Mean ± SD (n = 3). (B) Db10-Glyco-scTRAIL-FAVSGAA was tested in 3 concentrations for combined binding to the EGFR+ tumor cell lines Colo205 and HCT116 by flow cytometry. In addition, a 200-fold molar excess of cetuximab was used for competition with the scTRAIL fusion protein, demonstrating residual binding to TRAIL receptors mediated by scTRAIL in its dimeric configuration. Mean ± SD (n = 3). (C) The binding (ELISA) of Db10-Glyco-scTRAIL-FAVSGAA (filled circles) and Db10-Glyco-scTRAIL-FAVSGAA competed with 10 µM cetuximab (open circles) was tested on purified EGFR-Fc. In addition, a FLAG-tagged, fully humanized anti-EGFR IgG derived from cetuximab (M. Siegemund, R. Kontermann, unpublished data) was tested with (open squares) and without (filled squares) competition by cetuximab. Db10-Glyco-scTRAIL-FAVSGAA and αEGFR-IgG bound to EGFR-Fc with EC50 values of 284 ± 34 pM and 77 ± 8 pM, respectively. Mean ± SD (n = 3). (D) The binding (ELISA) of Db10-Glyco-scTRAIL-FAVSGAA (filled symbols) was tested on purified DR4-Fc and DR5-Fc in comparison to Db-Glyco-scTRAIL-95L8 (open symbols). Mean ± SD (n = 3).