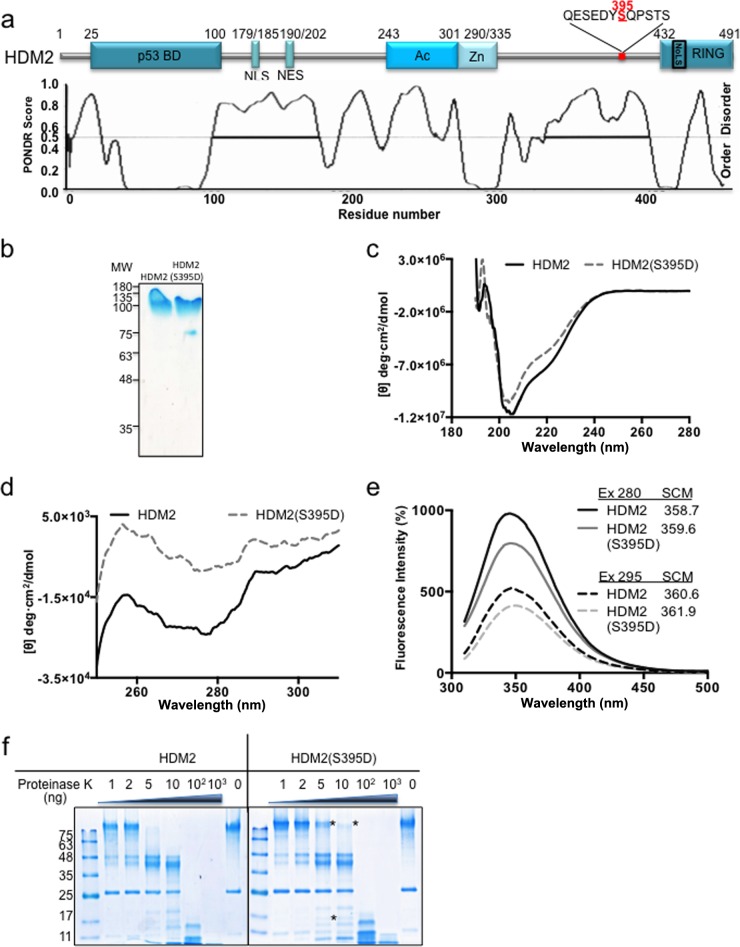
FIG 1.
HDM2 conformational rearrangement promoted by phosphomimetic mutant HDM2(S395D). (a) Cartoon illustrating major domains of HDM2. p53 BD, the p53 binding domain; NLS, nuclear localization signal; NES, nuclear export signal; Ac, acidic domain; Zn, zinc finger; RING, really interesting new gene domain. The predicted intrinsically disordered regions of HDM2 are shown in the lower panel. Serine 395 is indicated in red and is located at the end of a disordered region. (b) Recombinant purified HDM2 and HDM2(S395D). MW, molecular weights in thousands. (c) Far-UV CD spectra of the HDM2 and phosphomimetic mutant HDM2(S395D). (d) Near-UV CD spectra of the HDM2 and phosphomimetic mutant HDM2(S395D). (e) The intrinsic fluorescence emission spectra of HDM2 and phosphomimetic mutant HDM2(S395D). The spectral center of mass (SCM) at excitation (Ex) wavelengths of 280 and 295 nm were calculated. All spectra shown were obtained after subtracting the blank (no enzyme) from the experimental values. (f) Limited proteolysis of HDM2 and the phosphomimetic mutant S395D by proteinase K. Asterisks represent the main differences between the wild type and the mutant, showing that HDM2(S395D) is less susceptible to proteinase K. One representative experiment of 3 is shown.